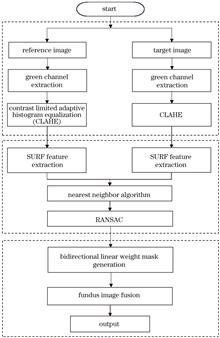
ObjectiveIn ophthalmic medical imaging, the stitching of ultra-wide-angle fundus images is essential for comprehensively observing and assessing patients' retinal health. This technology provides a broader visual field, allowing doctors to gain a more intuitive understanding of the entire fundus region, thereby offering crucial support for early screening, diagnosis, and treatment planning of diseases. However, stitching ultra-wide-angle fundus images faces multiple challenges, particularly in multi-scene and multi-angle shooting scenarios in practical applications. Variations in perspective and exposure differences can lead to significant issues in image matching and fusion, such as the formation of stitching seams. This phenomenon not only affects image quality but may also obscure the identification of lesion areas by doctors. To address these challenges, we aim to develop an efficient algorithm that ensures high-quality, seamless stitching under varying exposure conditions and angles. By minimizing stitching seams and enhancing image smoothness, we seek to provide more precise and reliable technical support for the diagnosis and monitoring of fundus diseases.MethodsWe present a novel image-stitching approach based on computer vision to address critical challenges in stitching ultra-wide-angle fundus images. The speeded-up robust features (SURF) algorithm was first employed to extract key feature points that accurately depict prominent regions, such as bifurcation points of retinal vessels and structural boundaries, from fundus images. Potential correspondences between feature points in different images were identified through initial matching. However, the initial matching outcomes may include a considerable number of mismatched points, affecting stitching accuracy. To refine the results, the random sample consensus (RANSAC) algorithm was applied after initial matching. Through an iterative approach, the RANSAC algorithm eliminates mismatched points and preserves true feature matches, ultimately deriving an accurate transformation matrix for geometric image registration. To address stitching seam issues caused by varying viewpoints and exposure differences, this study introduces an innovative bidirectional linear weight fusion method. This method followed a structured process. Firstly, the center point of the overlapping area was extracted, and an image rotation alignment technique was used to ensure correct geometric alignment of the images. Then, weights were assigned to overlapping areas, forming a bidirectional linear weight mask, enabling the pixel values in the transition area to be smoothly fused. Then, a mask with linearly decreasing weights was generated, ensuring a smooth transition between images and effectively eliminating the stitching seams caused by exposure variations.Results and DiscussionsThrough an experimental verification, the proposed stitching algorithm—combining SURF and bidirectional linear weight fusion—demonstrates significant performance advantages under various exposure conditions and viewing angles. Compared with the traditional algorithms, such as maximum fusion algorithm and gradual in and gradual out fusion algorithm, this algorithm achieves superior visual effects and smoother stitching. Experimental results show that this algorithm significantly reduces the stitching saliency when processing images with substantial exposure differences. This improvement is reflected as a 50.43% reduction in the average gradient and an 11.91% decrease in standard deviation in the stitching area compared to traditional algorithms, indicating a significant enhancement in seamlessness. Additionally, this algorithm effectively preserves image integrity. Information entropy, a key metric for measuring image information content, is only 3.13% lower than that of traditional algorithms. This finding suggests that while weight fusion eliminates stitching seams, the overall richness of image information remains nearly intact. As a result, the stitched images are not only more visually coherent but also retain critical medical details, providing reliable support for fundus disease diagnosis.ConclusionsBased on experimental results and theoretical analysis, we propose a fundus image stitching method that integrates SURF and bidirectional linear weight fusion, demonstrating excellent performance in addressing multi-scene and multi-angle stitching challenges. SURF extracts precise feature points, while the RANSAC algorithm ensures geometric registration, thereby enhancing stitching accuracy. To resolve exposure differences in overlapping areas, a bidirectional linear weight mask is designed for effectively eliminating stitching seams and significantly improving image smoothness and visual coherence. Experimental results further confirm that this algorithm outperforms traditional approaches in terms of mean gradient and standard deviation in stitched areas while also preserving information integrity. The slight 3.13% reduction in information entropy indicates that the method effectively balances seamless stitching with the retention of medical image details. This advancement is particularly valuable for medical diagnostic applications requiring high-precision panoramic fundus images, such as the early detection and monitoring of diabetic retinopathy and glaucoma. In conclusion, the proposed algorithm not only achieves high-precision, seamless stitching, but also introduces innovative tools and techniques in the field of ophthalmic medical imaging. These findings highlight its broad clinical applicability and potential for extension to more complex medical image analysis scenarios, fostering advancements in medical imaging technology.
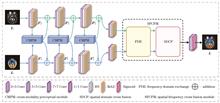
ObjectiveMedical image fusion integrates lesion features and complementary information from different modalities, offering a comprehensive and accurate description of medical images for clinical diagnosis. Traditional methods often result in reduced contrast and spectral degradation due to differences between multimodal images. Frequency-domain techniques mitigate these issues but rely on manually designed feature extraction and fusion rules, lacking robustness and adaptability. While deep learning-based fusion methods, such as convolutional neural networks and Transformer, have shown promising results in feature extraction and reconstruction, they often overlook complementary characteristics between modalities, leading to insufficient global information capture. Although frequency-domain methods preserve high-frequency information, they fail to adequately correlate global and local features, neglecting unique aspects of each modality and resulting in excessive smoothing and blurring. This study proposes an adaptive medical image fusion method based on cross-modality perception and spatial-frequency interaction.MethodsAn adaptive medical image fusion network combining cross-modality perception with spatial-frequency interaction was developed. First, a cross-modality perceptual module utilizing channel and coordinate attention mechanisms extracts multiscale deep features and local abnormality information, reducing information loss between modalities. Second, a spatial-frequency cross-fusion module based on frequency information exchange and spatial domain adaptive cross-fusion was proposed. This module alleviates information imbalance between modalities by exchanging phase information in the frequency domain and dynamically learning global interaction features in the spatial domain. This process highlights prominent targets while preserving critical pathological information and texture details. Finally, a loss function comprising content, structure, and spectral terms was designed to further improve the quality of the fused image.Results and DiscussionsFusion experiment for mild Alzheimer's disease demonstrates that the proposed method better preserves positron emission tomography functional information and edge details of magnetic resonance imaging (MRI) soft tissue, improving image contrast and detail presentation compared to other methods. The fusion experiment for metastatic bronchogenic carcinoma shows that other methods suffer from low resolution, blurred textures, and noise interference, hindering the observation and diagnosis of the lesion area. In contrast, the proposed method effectively retains single-photon emission computed tomography metabolic information and MRI soft tissue edge details, enabling doctors to comprehensively evaluate lesion status. The sarcoma fusion experiment further validates the algorithm’s effectiveness in preserving tissue edges, grayscale information, and density structure integrity. From tables 1?3, the AG (average gradient), MI (mutual information), SF (spatial frequency), QAB/F (fusion quality), CC (correlation coefficient), and VIF (visual information fidelity) indicators demonstrate strong performance. Specifically, the high MI value indicates that the fusion image contains rich features and edge information. The high SF value shows that the fusion image retains additional global information from the source images, with clear details and texture features. The high VIF value reflects consistency with the human eye’s visual characteristics, while the high QAB/F value indicates that the fusion image maintains spatial details consistent with the source images. Compared with other algorithms, the proposed method emphasizes the perception and interaction of structural image texture contours and functional image metabolic brightness during feature extraction and fusion, addressing issues such as structural edge loss and lesion detail blurring in existing fusion methods.ConclusionsTo enhance the quality of multimodal medical image fusion, this study proposes a method combining cross-modality perception with spatial-frequency interaction. During feature extraction, a multiscale cross-modality perception network facilitates the interaction of structural and functional information, fully extracting source image data and enhancing local lesion features. In the fusion stage, functional and anatomical key information is preserved through frequency-domain exchange, followed by cross-attention for adaptive fusion, ensuring that detailed texture and overall edge profile information are fully fused. Additionally, content, structural, and spectral losses were designed to retain complementary and chromatic information. Experimental results demonstrate that the proposed method improves AG, MI, SF, QAB/F, CC, and VIF by 4.4%, 13.2%, 2.7%, 3.4%, 11%, and 3%, respectively, showing that the method effectively retains unique information from each modality, resulting in fusion images with clear edges, rich lesion details, and high visual fidelity. In the task of fusing multimodal medical images of the abdomen with green fluorescent protein and phase contrast images, the proposed method demonstrates strong generalization, supporting its potential for application in other biomedical diagnostic tasks and enhancing clinicians’ diagnostic efficiency.
SignificanceThe optical attenuation coefficient (AC), a fundamental tissue parameter, quantifies the rate at which light diminishes as it propagates through a medium. This parameter is crucial for the quantitative analysis of tissue properties using optical coherence tomography (OCT) signals. Pathological changes in biological tissues induce considerable alterations in their complex morphological structures and optical properties, which are influenced by factors such as tissue composition, architecture, and physiological conditions. OCT technology, which measures the intensity and temporal delay of faint coherent reflections and backscattered light, enables rapid, noncontact, and high-resolution in vivo imaging of tissues. Quantitative OCT (qOCT) combines OCT with advanced algorithms to extract tissue optical properties. This technique provides detailed morphological insights and quantitatively evaluates AC, providing highly precise information on tissue morphology, composition, and lesion detection.This review explores the principles of tissue AC extraction using qOCT (Fig. 2). The review also provides a comprehensive summary of the algorithms used to extract tissue AC values via qOCT, and examines the advantages, limitations, and applicable scenarios of the selected commonly used algorithms. Finally, the clinical applications of qOCT for extracting tissue AC values are discussed, along with the associated challenges and potential future directions for development.ProgressThe theoretical foundation of the algorithms used in qOCT primarily encompasses single scattering (SS) and multiple scattering (MS) models (Fig. 3). The SS model, which is suitable for weakly scattering samples or thin layers of densely scattering tissues, assumes a single backscattering event. Prominent SS-based algorithms include curve-fitting, fast frequency-domain, and depth-resolved methods, each with distinct strengths and limitations. In contrast, the MS model accounts for multiple scattering events, requiring a detailed analysis of the photon propagation pathways and probability distributions within tissues. Although these models are based on complex physical frameworks and computational methods that offer higher precision, they typically come with a tradeoff of slower processing speeds. Common MS-based algorithms include the Monte Carlo method, the extended Huygens?Fresnel model, and the Maxwell’s equations?based method. In addition, our research group has introduced an innovative approach called the multi-reference phantom-driven network. This approach employs multi-reference phantoms and deep learning techniques to implicitly model factors influencing OCT signal propagation, thereby enabling automated and accurate regression of the AC.The accuracy of AC extraction is influenced by various factors, including the detection systems, signal acquisition protocols, and processing methodologies (Fig. 4). Hardware parameters of the OCT detection system, such as the light source specifications, probe design, and system type, are critical for ensuring reliable calculation of the AC values. Preprocessing steps, including noise reduction, contrast enhancement, artifact removal, and motion correction, are essential for achieving accurate AC computations. Moreover, tissue heterogeneity, multilayer structures, and pathological changes, such as cell aggregation, neovascularization, or fibrosis, can complicate light?tissue interactions and reduce the accuracy of AC calculations.qOCT offers high-resolution, quantitative insights into the optical properties of tissue and has demonstrated initial clinical applications in detecting ophthalmic, luminal, cancerous, superficial, and other diseases (Fig. 6). In ophthalmology, qOCT has become a vital tool for detecting and monitoring conditions such as glaucoma, macular degeneration, and diabetic retinopathy, enabling early intervention and improving patient outcomes (Fig. 7). In cancer diagnostics, qOCT can identify discernible changes in tissue morphology that result in notable alterations in AC and serve as a valuable biomarker for cancer monitoring and staging (Fig. 8). Moreover, qOCT has been increasingly recognized for its application in cardiovascular assessments, particularly in the detection and analysis of atherosclerotic plaques (Fig. 9). The ability of qOCT to precisely extract tissue AC values enables the quantitative assessment of lesions, offering robust support for the diagnosis and management of superficial conditions, such as skin lesions (Fig. 10). Additionally, qOCT holds great promise in the examination of lymphatic tissues, providing high-resolution images that reveal lymph nodes, vessels, blood vessels, and other microscopic structures, offering valuable insights into the structure and function of lymphatic tissues (Fig. 11).Conclusions and ProspectsAs an innovative quantitative analysis technique, qOCT enables the nondestructive, real-time acquisition of structural information and tissue AC in vivo, allowing for precise quantification of tissue structure and composition. The notable changes in tissue AC associated with pathological alterations provide robust clinical diagnostics using qOCT. Ongoing research focuses on developing depth-resolved, high-sensitivity, and high-resolution qOCT technologies for the real-time in vivo quantification of human tissues. These advancements aim to address existing challenges and broaden the clinical applications of this promising technology.
SignificanceCells are fundamental structural and functional units of all organisms. Visual tracking and analysis of intracellular processes are essential for understanding biological mechanisms, preventing disease, and developing therapeutic interventions. However, traditional optical microscopy is inherently limited by a diffraction barrier, which restricts its resolution to approximately 200 nm. This limitation poses significant challenges in resolving subcellular structures and observing dynamic biological processes at the nanoscale. To overcome these limitations, super-resolution microscopy techniques, such as stimulated emission depletion (STED), structured illumination microscopy (SIM), and photoactivated localization microscopy (PALM), have been developed. These technologies have successfully pushed the boundaries of optical resolution, unlocking new possibilities for nanoscale visualization. However, their practical applications are often constrained by challenges, such as high phototoxicity, complex system configurations, limited imaging depth, and time-consuming image processing. These issues have hindered their adoption in live-cell imaging and thick-sample studies.Recently, the introduction of lanthanide-doped upconversion nanoparticles (UCNPs) has provided innovative solutions for several of these challenges. UCNPs possess unique optical properties that enable them to convert near-infrared (NIR) light into higher energy emissions such as visible and ultraviolet light. This upconversion process offers several advantages: (1) NIR light enhances tissue penetration depth because of its low scattering and absorption in biological tissues; (2) the reduced energy of NIR excitation minimizes photodamage and phototoxicity, making UCNPs suitable for long-term live-cell imaging; (3) their high photostability mitigates photobleaching, enabling extended imaging sessions; and (4) their ability to produce multicolor emissions through precise doping strategies facilitates multiplexed imaging. These properties make UCNPs exceptional tools for advancing super-resolution microscopy, particularly in applications requiring deep tissue imaging and dynamic biological studies.ProgressThis review comprehensively discusses the integration of UCNPs into super-resolution microscopy techniques, highlighting their impact in overcoming the existing challenges and expanding their application potential. Key advances include(1) Stimulated emission depletion super-resolution microscopy based on UCNPs. The combination of UCNPs with STED microscopy leverages their nonlinear optical properties and photon avalanche effects to achieve resolutions below 30 nm. UCNPs-enhanced STED systems require a lower depletion laser power, reducing photodamage and enabling long-term imaging. These systems have been successfully applied to dual-color imaging and temperature-sensitive measurements, offering unprecedented precision in nanoscale visualization (Fig. 3).(2) Stimulated emission depletion-like super-resolution microscopy based on UCNPs. UCNPs provide a broader prospect for the development and enrichment of STED-like super-resolution imaging by significantly simplifying the complexity of imaging systems, significantly reducing the excitation power, and improving the imaging resolution. This lays the theoretical foundation for the proposal and progress of Fourier-domain heterochromatic fusion and near-infrared emission saturation microscopy. Simultaneously, UCNPs can be integrated with current microscopy systems, and the current hardware and algorithms can be continuously optimized, which is expected to achieve instant subtraction. This in turn enables real-time, low-power super-resolution microscopy and promotes major breakthroughs in fields such as biomedical research and materials science (Figs. 5 and 6).(3) Structured illumination microscopy based on UCNPs. UCNPs provide more nonlinear information for SIM super-resolution imaging. This in turn effectively improves the resolution of SIM. Specifically, the dual near-infrared light characteristics of near-infrared excitation and fluorescence emission enhance SIM suitability for biological tissue imaging by increasing biological penetration. Combined with the fluorescence lifetime characteristics of UCNPs, SIM highlights its multi-channel imaging capabilities, achieving higher decoding accuracy and high-throughput photomultiplexing imaging efficiency. In the future, by further enhancing the emission intensity of UCNPs and optimizing imaging equipment, the performance of U-SIM is expected to improve further, promoting its application in biomedical imaging (Fig. 8).(4) Super-linear and photon avalanche mechanisms. Photon avalanche effects in UCNPs amplify nonlinear responses, enabling ultrahigh-resolution imaging with minimal system modifications. Super-linear excitation-emission (SEE) microscopy takes advantage of these high-order nonlinear effects, achieving a resolution of 62 nm. These approaches simplify the integration of UCNPs into existing imaging systems while maintaining high precision (Fig. 9).(5) Other super-resolution techniques. UCNPs have been integrated into cutting-edge imaging methods, including single-molecule localization microscopy (SMLM) and nanoscale optical writing. Their reversible photophysical properties facilitate dynamic imaging and nanoscale patterning. Furthermore, UCNPs show promise in emerging applications such as temperature-sensitive imaging, multiplexed fluorescence detection, and functionalized imaging for specific biomolecular interactions (Fig. 10).Conclusions and ProspectsUCNPs represent a transformative leap in super-resolution microscopy, addressing key challenges such as phototoxicity, limited imaging depth, and system complexity. By enabling deeper tissue imaging, reducing photodamage, and achieving higher resolution, UCNPs are poised to revolutionize biomedical imaging and material sciences. Future research should focus on optimizing the UCNPs design, enhancing the emission intensity, and developing advanced computational algorithms to fully unlock their potential. Improving the scalability and integration of UCNPs with existing imaging systems is critical for their broad adoption in real-time, low-power, and high-resolution applications. These advancements promise the establishment of UCNPs as indispensable tools for next-generation imaging technologies and drive innovations in biomedical research, diagnostics, and materials science.
SignificanceThe macular fovea in the retina has the highest density of photoreceptor cells making it the region with the sharpest visual acuity. It contains the only deep capillary network in the human body that can be directly observed using optical imaging methods. Obtaining high-resolution imagery of the deep capillary network and functional assessment of blood flow are of paramount importance for early diagnosis of retinal diseases. With the integration of adaptive optics technology and various retinal imaging techniques, such as flood illumination ophthalmoscopy, confocal scanning laser ophthalmoscopy, and optical coherence tomography, photoreceptors and retinal capillaries can be clearly distinguished, allowing precise measurement of the blood flow velocity in the capillaries. This advancement offers new perspectives and insights into the diagnosis and research of retinal diseases. This study explores the applications and recent advancements of adaptive optics single-cell-resolution retinal imaging technology, specifically in retinal capillary imaging and blood flow measurement. Further, it analyzes existing difficulties and technical challenges and summarizes strategies to overcome these challenges and future development prospects.ObjectiveOptical imaging is the most prevalent, well-established, and reliable method for observing retinal blood vessels. The most commonly used ophthalmic optical imaging devices in clinical practice currently, such as fundus cameras and confocal laser scanning ophthalmoscopes, can directly collect retinal reflected light to generate retinal reflection images. Moreover, they can produce high-contrast images of the retinal vasculature when combined with fluorescein angiography. Optical coherence tomography (OCT) is based on the principle of low-coherence light interference and enables three-dimensional imaging of retinal tissue structures. When combined with Doppler imaging techniques, OCT can provide additional real-time data on the blood flow velocity within the retinal vessels. However, owing to the influence of ocular aberrations, these methods can only resolve retinal vessels with a diameter greater than 30 μm. By the time these blood vessels begin to exhibit pathological features, the retinal tissue structures and functions may have already undergone irreversible damage. Consequently, treatment at this stage is mostly poor compared to interventions initiated during the early stages of the disease.Adaptive optics technology has been integrated into retinal imaging techniques to improve spatial resolution. By correcting the dynamic aberrations produced by the living human eye, which vary in both time and space, it is possible to achieve imaging resolutions close to the diffraction limit of the human eye, thus enabling retinal imaging at the single-cell-resolution level. With advancements in technology, adaptive optics has been successfully integrated with imaging techniques, such as flood illumination ophthalmoscopy, confocal laser scanning ophthalmoscopy, and optical coherence tomography, to obtain high-resolution images of various cells and tissue structures in the living human retina. Combined with the previously mentioned methods for detecting blood flow in retinal capillaries at single-cell-resolution, these techniques enable the observation and investigation of hemodynamic changes associated with diseases at an early stage.ProgressThe first adaptive optics flood illumination ophthalmoscope/fundus camera (AOFIO/AOFC) was built and commissioned in the United States in 1997. This was the result of the pioneering effort of the Center for Visual Science at the University of Rochester in the United States, towards the integration of the adaptive optics technology with flood illumination ophthalmoscopy. Consequently, detection and correction of higher-order aberrations in the human eye and obtaining high-resolution images of living human retinal cells became a reality for the first time in the world. Subsequently, adaptive optics technology was integrated with various retinal imaging techniques, including confocal scanning laser ophthalmoscopy, optical coherence tomography, and line scanning ophthalmoscopy. This led to the development of several adaptive optics ophthalmic imaging systems, such as the adaptive optics scanning laser ophthalmoscope (AOSLO), adaptive optics optical coherence tomography (AOOCT), and adaptive optics line scanning ophthalmoscope (AOLSO). These advancements have enabled single-cell-resolution imaging of retinal structures, including photoreceptor cells, the retinal pigment epithelium, retinal capillaries, and retinal nerve fibers. Consequently, it is possible to perform retinal hemodynamic assessments at the cellular level. In the early detection of diseases such as diabetic retinopathy, age-related macular degeneration, Alzheimer’s disease, and hypertension, adaptive optics ophthalmic imaging systems have successfully observed structural and physiological changes in retinal capillaries caused by these diseases, thereby demonstrating their potential for clinical applications.Conclusions and ProspectsThe introduction of adaptive optics technology has enabled in vivo optical imaging of the human retina at single-cell-resolution, leading to significant advancements in retinal research for both healthy and diseased eyes. To advance clinical application of single-cell-resolution retinal imaging technology and single-cell-resolution retinal capillary blood flow detection, future research and development will focus on the key areas listed hereunder.1) Substantial expansion of the field-of-view for single-cell-resolution imaging, which is bound to be a remarkable advancement in the development of high-resolution imaging. Technologies such as multi-conjugate adaptive optics, multi-pupil imaging, and rapid image montaging are dedicated to achieving large field-of-view in single-cell-resolution retinal imaging.2) Expanding the integration of single-cell-resolution imaging systems through system size reduction, simplification of operational procedures, development of more straightforward methods for wavefront aberration measurement and closed-loop correction, and reduction of hardware costs and operational complexity.3) Developing more accurate and faster image processing and analysis softwares to clarify the efficacy, reproducibility, and application scope of single-cell-resolution imaging in clinical diagnostics. Ultimately, this will result in the establishment of standardized clinical detection methods and criteria. With continuous technological advancements, single-cell retinal imaging is expected to find wider applications in both clinical practice and scientific research, thereby establishing a new standard for high-resolution single-cell diagnosis and treatment. This development will facilitate a deeper understanding of the pathogenesis of various ophthalmic diseases, clarify the relationship between visual perception and retinal structure, enhance the accuracy and effectiveness of patient treatment, and enable the early diagnosis and treatment of ocular diseases.
SignificanceThe kidney, a vital organ in the human body, is responsible for filtering blood, removing waste and excess water, and precisely regulating electrolyte balance and acid?base levels. Additionally, it plays a crucial role in controlling blood pressure, stimulating red blood cell production, and activating vitamin D. However, acute or chronic kidney injuries can lead to physiological changes that impair these normal functions. Existing clinical assessment methods have limitations in detecting kidney health status, including the inability to provide real-time monitoring, invasiveness, high cost, and low image resolution. Optical coherence tomography (OCT), with its advantages of high resolution, rapid imaging, and non-invasiveness, holds considerable promise for assessing kidney health and diagnosing related diseases.ProgressWe review the latest advancements in OCT for kidney detection, focusing on the technological development of OCT in kidney research, its application in imaging kidney microstructure and microcirculation, its clinical applications, its role in providing comprehensive optical characterization of the kidney, and the use of large-field OCT for kidney assessment. First, we introduce the application of OCT imaging in assessing kidney microstructure and microcirculation. Since 2007, OCT has been widely used to study the microstructure of murine kidneys, yielding results consistent with histopathological findings (Fig. 1). Renal imaging with OCT enables real-time monitoring of morphological changes in transplanted kidneys and provides quantitative information through image analysis, aiding in the assessment of renal function and acute tubular injury. Vascular imaging techniques such as Doppler OCT, OCT angiography (OCTA), and optical microangiography (OCMA) enhance the ability to monitor kidney microcirculation and accurately quantify blood flow (Fig. 2). Studies have demonstrated the effectiveness of OCMA in monitoring the effects of drug delivery systems on renal blood circulation. Previous research has shown that OCT can effectively penetrate the connective tissue surrounding the human kidney, enabling non-destructive imaging of the renal cortex and identification of histopathological changes in kidney microstructure (Fig. 3). Machine learning algorithms have been employed to automatically identify and segment kidney microstructures in OCT images, achieving accuracy comparable to manual segmentation while eliminating subjective bias. Additionally, OCT can distinguish between renal tumors and normal renal parenchyma using attenuation coefficient measurements, enabling accurate tumor characterization (Fig. 4). These findings suggest that OCT is a safe and reliable technique for real-time observation of donor renal structures during transplantation, providing valuable quantitative data for predicting post-transplant renal function recovery. Advances in automated scanning technology and artificial intelligence (AI)-based image analysis have further simplified OCT usage, rendering it more accessible and precise.Next, we explore the expanding role of OCT in comprehensive optical characterization of the kidney. Polarization-sensitive OCT (PS-OCT) has demonstrated a strong ability to accurately identify tumor regions at various depths and locations, as well as to reconstruct tumor structures in 3D (Fig. 5). The combination of OCT with optical coherence elastography (OCE) has proven valuable for identifying and classifying nephritis. Morphological differences in renal tubule functional activity can be effectively visualized using dynamic OCT (Fig. 6). These studies highlight the potential clinical applications of OCT in diagnosing and evaluating multiple kidney conditions in humans. Advancements in imaging technology have further enhanced OCT’s clinical utility. Handheld imaging probe positioning systems and robotic arm-assisted OCT represent significant developments in the field. The handheld imaging probe positioning system can precisely locate and reconstruct images based on the OCT probe’s position, as determined by visual odometry (VO) (Fig. 7). Robotic arm-assisted automated scanning streamlines medical workflows, accelerating the acquisition of objective quantitative analysis results (Fig. 8). Although robotic arms enable automatic, high-precision, and large-field scanning, further testing, adjustments, and optimization in clinical settings are necessary to improve adaptability to medical applications. Researchers have also investigated the feasibility of needle-based OCT for renal tumor assessment, concluding that it offers real-time data acquisition, improved tumor localization, and enhanced diagnostic accuracy (Fig. 9). Additionally, the integration of OCT endoscopy with deep learning has yielded remarkable results in the automatic classification of OCT renal images, significantly improving diagnostic efficiency.Finally, we introduce studies exploring the correlation between fundus OCT blood vessel images and kidney injury. Given that renal microvascular changes play a key role in chronic kidney disease (CKD) and that renal biopsy has limitations in assessing these changes longitudinally and in real time, the visualization of the inner retina as a surrogate marker for kidney disease progression presents a novel avenue for noninvasive diagnosis. Retinal and choroidal changes observed using OCT, as potential indicators of renal pathology, have garnered significant interest. The potential ability of OCT-derived parameters from retinal and choroidal imaging to detect and monitor intraocular vascular injury, as well as their feasibility as alternative biomarkers for renal vascular injury, offers a promising approach for simplifying and enhancing the noninvasive diagnosis of kidney diseases. This novel strategy provides a safer and more effective means of identifying renal abnormalities without inconveniencing the patients.Conclusions and ProspectsOCT has been widely utilized in nephrology due to its noninvasiveness, high resolution, and real-time imaging. Although minimally invasive imaging techniques and fundus-based correlation studies are still in their early stages, they offer a progressive diagnostic pathway—from noninvasive retinal and choroidal examinations to minimally invasive imaging, and ultimately, if necessary, to surgical intervention. This hierarchical diagnostic strategy is expected to improve the efficiency and accuracy of kidney disease diagnosis and support the advancement of precision medicine in clinical nephrology.
SignificanceIn recent decades, light-sheet fluorescence microscopy (LSFM) has emerged as a groundbreaking technique in the field of fluorescence microscopy, offering unparalleled tomographic capabilities. Using thin light sheets for imaging, LSFM dramatically reduces phototoxicity and photo-bleaching, enabling researchers to capture high-quality and real-time images of living specimens over extended periods without compromising the integrity of the sample. This noninvasive approach has revolutionized the visualization and quantification of biological processes, providing insight into developmental biology, cellular dynamics, and disease mechanisms. As research in the life sciences continues to expand, the demand for more sophisticated imaging technologies capable of capturing complex biological phenomena across spatial and temporal scales has increased. Despite the advantages of LSFM, several challenges remain in overcoming traditional imaging trade-offs, such as spatial resolution, temporal resolution, field-of-view, and the health of the sample. To address these issues, the integration of artificial intelligence (AI) and deep learning algorithms has become a focal point for enhancing the capabilities of LSFM and paving the way for the next generation of intelligent imaging systems.ProgressThis review summarizes the recent advances in LSFM, particularly in the integration of AI-driven intelligent imaging systems and image restoration technologies. Recent developments in smart adaptive imaging schemes (Fig. 3), powered by deep learning techniques, represent a transformative role in overcoming the inherent limitations of traditional microscopies. These methods enable real-time adjustments to imaging conditions, ensuring that images are captured at optimal resolution and contrast, while minimizing phototoxicity and preserving sample integrity. In particular, AI algorithms enable the automatic selection of imaging parameters based on sample characteristics, thus enhancing the efficiency and accuracy of data acquisition.Significant progress has also been made in developing image restoration techniques that enhance the quality of LSFM images, even under challenging conditions, such as low signal-to-noise ratios or large-scale imaging (Fig. 4). By applying deep learning models to noise reduction, resolution enhancement, and artifact removal, researchers can now achieve high-fidelity reconstructions of biological structures, even when confronted with imperfect or incomplete data. These advancements in image processing are important for unlocking the full potential of LSFM in complex biological studies.This review also highlights the role of AI in optimizing data processing workflows (Fig. 5). The enormous volume of data generated by LSFM, particularly in multidimensional imaging, necessitates efficient algorithms for data management, analysis, and visualization. Machine learning algorithms, particularly convolutional neural networks and other deep learning frameworks, have been instrumental in automating the interpretation of large-scale datasets, which has facilitated the extraction of meaningful biological insights with minimal manual intervention.The integration of AI and deep learning into LSFM has already demonstrated marked improvements in a wide range of applications, including live-cell imaging, developmental biology, neuroimaging, and cancer research. These innovations not only extend the capabilities of LSFM, but provide life scientists with powerful tools to explore biological processes in unprecedented detail and over longer time frames.Conclusions and ProspectsIn the future, the development of LSFM is poised to be shaped by continued advancements in AI and machine learning. The next phase of LSFM innovation will focus on further enhancing spatial and temporal resolution, increasing imaging throughput, and improving multiscale imaging capabilities. AI will likely continue to play a major role in refining image restoration techniques and streamlining data processing workflows, thus enabling the capture of more accurate and informative biological images.In particular, the integration of LSFM with complementary imaging technologies, such as super-resolution microscopy and multi-photon microscopy, holds promise for achieving more comprehensive, high-resolution imaging at the molecular and cellular level. As these technologies advance, they will offer unprecedented insight into the molecular mechanisms underlying various diseases, including cancer and neurodegenerative disorders, and open up new possibilities for drug discovery and personalized medicine.As LSFM systems become automated, the need for standardized protocols and best practices is also growing to ensure consistency and reproducibility across diverse research environments. Efforts to establish universal standards for AI-driven LSFM systems will be necessary for facilitating the widespread adoption of these technologies in research and clinical settings. Ultimately, the convergence of AI, deep learning, and LSFM will unlock new frontiers in biomedical research, enabling the next generation of discoveries in the life sciences.
ObjectiveIn contemporary society, dental diseases affect people of all ages, increasing the workload of dentists. Oral panoramic imaging is a widely used diagnostic tool in dentistry, and doctors must process image data from numerous patients amid their heavy daily clinical workload. However, manually analyzing complex image data is time-consuming, laborious, and susceptible to various human factors, such as fatigue, emotional fluctuations, and differences in professional skills. These factors can adversely affect diagnostic accuracy, delay treatment, and damage patient health. Although artificial intelligence (AI) is initially applied in dental disease detection, most current AI research focuses on single disease or restoration. However, when the number of detection targets increases, the decrease in detection accuracy can hinder practical clinical applications. Therefore, this study applies deep learning to identify key image features for efficient and accurate lesion screening in oral panoramic images using a deep learning network architecture. The purpose is to detect abnormal teeth and restorations including dental cavities, blocked teeth, implants, root canal treated teeth, fillings, crowns, and bridges. Intelligent assistance methods can be used to reduce human errors, accelerate diagnoses, and improve medical quality and efficiency.MethodsThis study proposes an intelligent assisted diagnostic network based on the YOLOv8 framework, designing a YOLOv8 model specifically for dental imaging. The purpose is to detect abnormal teeth and restorations including cavities, residual teeth, implants, root canal treated teeth, fillings, crowns, and bridges. Intelligent assistance methods can be used to reduce human error, accelerate diagnosis, and improve medical quality and efficiency. First, to enhance feature extraction capability, we integrated a spatial grouping enhancement (SGE) attention mechanism to enhance the model ability to capture complex oral features. In addition, to address the difficulty of identifying small lesions, a small-object detection layer was added. This layer integrates multiple features and maintains detailed information, thereby enhancing the capability of the model in detecting fine lesions. Subsequently, the model loss function was optimized, adopting the generalized intersection over union (GIoU) loss function to improve the prediction accuracy of bounding box, which further enhanced localization performance. Finally, to reduce the computational burden of improved model, the layer-adaptive magnitude-based pruning (LAMP) method was used. This method eliminates non-contributing channels and improves detection speed.Results and DiscussionsThe analysis in Table 2 shows that the SGE attention mechanism performs well in target recognition, outperforming other attention mechanisms in all detection results. Table 4 shows the results of the ablation experiment, indicating that integrating the SGE attention mechanism into the baseline model improves accuracy, recall, and mean average precision (mAP) by 2.4, 2.6, and 1.0 percentage points, respectively. This indicates that the SGE attention mechanism can effectively group features, improve recognition rate, enhance feature extraction, and suppress information interference. After the addition of small-object detection layer, accuracy, recall, and mAP increased by 3.0, 2.4, and 2.1 percentage points, respectively, indicating that the small-object detection layer effectively identifies smaller detection targets and enhances the network ability to recognize small objects. After replacing completing intersection over union (CIoU) with GIoU, the accuracy and mAP increased by 3.6 and 1.2 percentage points, respectively; however, the recall rate decreased by 0.7 percentage points. This indicates that GIoU enhances localization performance and improves recognition accuracy. The final model, YOLOv8-Dental, was developed using the LAMP method, which improved accuracy, recall, and mAP by 5.2, 4.6, and 5.2 percentage points, respectively, while reducing parameters and computational complexity by 2.02×106 and 0.9×109, respectively. Table 5 shows the comparative experiments, indicating that although YOLOv8-Dental performed slightly worse than some models in terms of implants and dental bridges, it still achieved recognition rates of 95.1% and 96.2% for these, respectively. In detecting the remaining five lesions, the proposed model outperformed the other models in average precision (AP) with fewer parameters and a lower computational workload. This ensures high detection accuracy for multiple lesions and maintains the overall detection rate.ConclusionsThis study explored the deep learning-based AI-assisted diagnosis of dental panoramas, aiming to reduce the healthcare burden of dentistry, assist dentists beyond the limitations of subjective judgment, and improve diagnostic accuracy. First, YOLOv8 was used as the base network, which was enhanced by integrating the SGE attention mechanism into its backbone feature extraction network. Second, to detect small target lesions in oral images, a small target detection layer was added to improve recognition accuracy. To further enhance the model bounding box localization accuracy, the GIoU loss function was adopted, which significantly improved the network bounding box regression performance. Finally, the model was pruned using the LAMP method to reduce the number of parameters and computation, thereby improving detection speed. All these optimization strategies were integrated to build the YOLOv8-Dental-assisted diagnosis model. Comparisons and ablation experiments demonstrated the positive impact of each optimization strategy on the diagnosis model. The experimental results showed that the YOLOv8-Dental model achieved a precision rate of 83.9%, recall rate of 87.8%, mAP of 89.8%, and frame rate of 409 frame/s for detecting cavities, blocked teeth, implants, root canal treated teeth, fillings, crowns, and bridges. The validity of the model was verified through physical image detection and heatmap analysis. The results of this study provide theoretical guidance and a methodological reference for deep-learning-based clinical diagnosis, promoting the research on deep-learning-based image-assisted diagnosis of dental diseases.
ObjectiveBrain tumor is a highly lethal cancer that occurs in human brain tissue, with glioma being one of the most prevalent types originating from glial cells. Malignant brain tumors can damage normal brain tissue and constrict key neural pathways, which may lead to symptoms such as headaches, seizures, vision loss, and limb weakness, significantly affecting the quality of life of patients. Therefore, early detection and treatment are crucial for managing the patient’s condition. Traditional manual segmentation methods are time-consuming, labor-intensive, and require professional knowledge. In recent years, due to the superiority of the convolutional neural network (CNN) in image feature extraction, it has rapidly gained attention in medical imaging. Classic models such as U-Net and V-Net perform well in capturing local and global features and processing three-dimensional (3D) data but exhibit significant computational complexity. Improved models enhance segmentation accuracy through attention mechanisms and hybrid architectures but frequently present challenges such as high memory consumption and slow training speed. Lightweight networks significantly reduce computational costs by optimizing the convolutional structures and reducing the parameter quantity, making them suitable for resource-constrained scenarios. However, there are shortcomings in the segmentation details and contextual modeling. Therefore, research on improving brain tumor segmentation networks is crucial for achieving intelligent healthcare with enormous clinical application potential. Given the above reasons, semiautomatic or fully automatic methods for brain tumor segmentation are being actively developed.MethodsThis study proposes a lightweight brain tumor segmentation network that balances global and local information, for high-precision segmentation performance with reduced parameters. This network is based on the U-Net architecture and introduces a semantic flow feature alignment mechanism to replace traditional skip connections. By learning the flow field, the feature maps of the encoder and decoder are spatially aligned to preserve semantic information and spatial details during the fusion process. In the feature extraction stage, the network adopts layered decoupled convolution units as the basic module while introducing shallow-scale perception modules as auxiliary branches to integrate multi-scale contextual information and facilitate adaptive adjustment of features. The scale perception module comprises two parts: multi-head mixed convolution and scale perception aggregation. The multi-head hybrid convolution combines the multi-head attention mechanism with multiscale residual convolution operation, effectively combining the global modeling ability of self-attention with the local feature extraction ability of the convolutional network. Scale-aware aggregation dynamically fuses multiscale features and adaptively modulates attention toward large-scale or intricate information according to regional attributes, thereby producing more discriminative feature representations. In deep feature extraction, the improved hierarchical decoupling convolution unit combined with multiscale convolution operation further enhances the feature capture capability while maintaining low computational complexity.Results and DiscussionsWe performed comparative experiments with other networks on the BraTS2020 dataset and generalization experiments on the BraTS2018 and BraTS2019 datasets. In the BraTS2020 dataset, unlike classical networks (Table 3), our network shows significantly higher Dice index in whole tumor (ET), tunor core (WT), and enhancing tunor (TC) areas than the other two networks. The 95% Hausdorff distance is significantly lower than the other two networks. The comparison of our model with the four popular networks used for brain tumor segmentation show that, the dResUnet model demonstrates the highest accuracy in the Dice index on ET area, the SwinBTS model shows the lowest accuracy, and the proposed method has a moderate effect compared to others. On WT area, the Dice index of SAHNet is 0.36 percentage points higher than the average accuracy, whereas, on TC area, it is about 2.83 percentage points higher than SwinBTS and about 1.08 percentage points lower than ASTNet. The overall effect of our network is at a medium to high level, and regarding the parameter quantity, it is considerably lesser than the other four networks. For lightweight networks, the segmentation performance of our network is superior to the other four segmentation methods. Compared to AD-Net, ET area shows an increase of about 1.12 percentage points in Dice index, while WT area remains the same accuracy. The Dice index in TC area is increased by 2.60 percentage points, and the number of parameters is decreased dramatically. Compared with the DMF network, our model outperforms in three indicators and has a much smaller number of parameters. Compared with HDC network, the number of parameters of our network is greater, whereas the Dice index shows increases of 0.43, 0.36, and 1.92 percentage points in ET, WT, and TC areas, respectively. In ET and WT areas, Dice index of our network is slightly lower than that of HMNet, whereas the accuracy in TC area exceeds that of HMNet by 1.93 percentage points. This indicates that the segmentation performance of the proposed network in TC area is much higher than that of other lightweight networks, while in WT and TC areas are equal. Our network emphasizes on detailed information without sacrificing other accuracies, and the segmentation effect is more accurate than other networks. The average segmentation accuracy of our network achieved on the BraTS2018 and BraTS2019 datasets reached 85.83% and 83.76%, respectively. Our model demonstrates good generalization ability compared with other lightweight segmentation methods (Table 4).ConclusionsThe SAHNet model proposed in this study adopts a layered decoupled convolution module as the basic feature extraction module. Compared with the traditional convolutions, it is more lightweight while maintaining a certain level of accuracy. Simultaneously, the hierarchical decoupling convolution is improved by proposing multi-scale hierarchical decoupling convolution to enhance the expressive power of the model. The feature alignment module is enhanced through the guidance of semantic flow and applied to skip connections, effectively improving the ability of feature alignment by generating flow fields and spatially distorting features. The scale perception module expands the receptive field of the convolution through local residual convolution in MHXC, enabling it to capture richer contextual information at different scales while preserving local features. The SAA module divides feature information into multiple groups and performs cross-group information fusion through lightweight 1×1×1 convolution to achieve global information crossover. The experiments using BraTS2018, BraTS2019, and BraTS2020 show that our method not only outperforms other lightweight networks in segmentation accuracy but also offers better deployment potential on resource-limited devices due to its lightweight design, which is expected to provide more efficient solutions for practical clinical applications.
ObjectiveMedical image registration is essential for surgical guidance and lesion monitoring. However, existing deep learning-based registration models typically rely on a single architecture, which limits the ability to leverage the complementary strengths of convolutional neural networks and Transformer models. This often leads to suboptimal registration accuracy and difficulties in preserving the original image topology. To address these challenges, a large kernel multi-scale convolution and Transformer-based parallel registration model (PLKCT-UNet) is proposed.MethodsWe develop PLKCT-UNet, a three dimensional (3D) medical image registration model that integrates large kernel convolution and Transformer parallel architecture. In the encoder, the model incorporates three key components. First, a large kernel multi-scale convolution module is designed to enhance the extraction of local detail information and manage large deformations effectively. Second, a 3D Swin Transformer module improves the model's capability to capture long-range dependencies, thereby enhancing generalization performance. Finally, a multi-scale attention aggregation strategy is employed to refine features after dual-encoder channel fusion, further boosting registration accuracy.Results and DiscussionsTo verify the effectiveness of the PLKCT-UNet model, experiments were conducted using the OASIS and LPBA40 datasets. In the comparative experiments, the OASIS dataset was utilized to calculate the degree of overlap between the segmentation masks of the moving and fixed images after registration using seven different methods and the proposed method.Results demonstrate that the proposed algorithm significantly improves registration performance while preserving the integrity of brain structures and maintaining local and spatial information. The algorithm achieves superior registration accuracy and maintains the continuity and consistency of anatomical structures, even under complex deformations. In the ablation experiments, the OASIS dataset was used to assess the contributions of the large kernel convolution (LKC) module, 3D Swin Transformer, and multi-scale attention aggregation (MSAA) module in medical image processing. Results indicate that each module contributes to enhancing the overall network performance. Generalizability experiments were performed using the LPBA40 dataset to validate the robustness of PLKCT-UNet across different datasets. Comparisons with six mainstream algorithms show that PLKCT-UNet achieves higher registration accuracy and generates smoother deformation fields, thereby improving the overall registration quality. These experiments confirm the stability and generalization capability of PLKCT-UNet, highlighting its significant advantages in handling complex deformations.ConclusionsThis study presents PLKCT-UNet, a novel registration model based on LKC and Transformer parallelism. The LKC module addresses sensory field size limitations, balancing detailed and global structures while employing kernel decomposition to reduce computational costs. The Swin Transformer module effectively captures long-range dependencies, enhancing the model's generalization ability. The MSAA module refines spatial and channel features through an attention aggregation strategy, improving dual-encoder feature fusion. On the OASIS dataset, the proposed model demonstrates superior registration performance compared to mainstream methods. Generalization experiments on the LPBA40 dataset further confirm its robustness and versatility. These results establish PLKCT-UNet as a state-of-the-art solution for unimodal medical image registration with broad application potential. Future work will focus on extending the algorithm to multimodal medical image registration and exploring more efficient optimization schemes to further enhance its practicality.
ObjectiveHigh-precision preoperative and intraoperative 3D point cloud registration during pedicle screw placement surgery is crucial for improving surgical safety and success rates. However, preoperative and intraoperative point clouds are obtained using different imaging devices and acquisition techniques, which give rise to challenges concerning noise, variations in densities, and initial poses of the two point clouds. In addition, the independent nature of keypoint features within the point cloud after encoding leads to a lack of global contextual correlation. The absence of feature interaction between keypoints of the preoperative and intraoperative point clouds further reduces the relevance of the features, resulting in suboptimal registration accuracy. To address these issues in preoperative and intraoperative point cloud registration for pedicle screw placement navigation systems and then improve the robustness and accuracy of the registration task, a cross-source point cloud registration network with enhanced attention mechanisms is proposed.MethodsA convolutional neural network is presented for preoperative and intraoperative point cloud registration with enhanced attention mechanisms. First, a voxel-filtering algorithm is applied to adjust the density of the intraoperative point cloud based on the density of the preoperative point cloud. Next, farthest point sampling (FPS) is employed to construct local regions of the preoperative point cloud. For local feature extraction, a multilayer perceptron (MLP) is used to build the encoder. Three feature extraction (FE) and feature propagation (FP) layers are employed to encode the point cloud into keypoints and their corresponding high-dimensional feature representations. The feature aggregation module, consisting of graph self-attention and cross-attention mechanisms, is used to enhance the feature representation of the point clouds. In the graph self-attention mechanism, K-nearest neighbors (KNN) is employed to connect each keypoint in the preoperative point cloud to its neighboring points. By calculating the differences between the keypoint features and neighboring point features, the expression of local geometric features is enhanced. The cross-attention mechanism captures the similarity between preoperative and intraoperative point clouds and identifies deep-level correlations to strengthen the global relevance. Then, the features obtained from cross-attention are enhanced using the graph self-attention mechanism to further improve the local contextual relationships. A similarity function is used to compute point-cloud-matching probabilities to obtain a set of corresponding keypoint pairs. Finally, the random sample consensus (RANSAC) algorithm is applied to eliminate incorrectly matched keypoint pairs. The accuracy of the calculated transformation matrix is improved.Results and DiscussionsTo verify the registration performance of the proposed cross-source point-cloud registration network with enhanced attention mechanisms in surgical navigation of pedicle screw placement, the following actions were conducted: algorithm comparisons, ablation experiments, and registration experiments on noise influenced by intraoperative data. The experiments were conducted on preoperative and intraoperative point cloud datasets, which comprise data from the Capital Medical University Affiliated Hospital and SpineWeb dataset, both of which exhibit substantial initial pose variation and angular changes. (Table 1). The proposed model successfully completes precise preoperative and intraoperative point cloud registration (Fig. 5). To evaluate the performance of the algorithm, the FPS+FPFH and FPS+FastReg methods are compared. The results (Table 2) demonstrate that the proposed method achieves the lowest error in coarse registration, with an average rotational error of 2.87° and a translational error of 3.22 mm, meeting clinical accuracy requirements. Additionally, to further analyze the impact of different attention mechanisms on the overall registration performance, ablation studies were designed to quantitatively assess the contributions of each module to the performance of the network. The results (Table 4) indicate that the combined use of graph self-attention and cross-attention mechanisms significantly improves the expression of point-cloud features and registration accuracy. Noise experiments were conducted to validate the robustness of the proposed model. The results (Table 5) show that although noise degrades performance, the proposed method still achieves good coarse registration accuracy under noisy conditions, demonstrating the robustness of the model to noise interference.ConclusionsTo address the challenges of significant initial poses and density differences during point-cloud registration in a pedicle screw placement navigation system, graph self-attention and cross-attention mechanisms are employed to aggregate and enhance features generated by the encoder. Graph self-attention refines the local feature representation, whereas cross-attention strengthens the global correlations between preoperative and intraoperative point clouds. Consequently, the integration of attention mechanisms allows the model to effectively capture the geometric structure of point clouds, improving both registration accuracy and robustness. The experimental results of the preoperative and intraoperative point cloud registration show that the proposed algorithm improves registration accuracy and efficiency in the navigation system for pedicle screw placement, even in cases with large initial pose differences and cross-source data. In addition, the proposed model demonstrates good coarse registration accuracy under noisy intraoperative point clouds, verifying its robustness against noise interference. Compared to the FPS+FPFH method and FPS+FastReg network model, the proposed model achieves better coarse registration accuracy with shorter executions. This algorithm improves the success rate of point-cloud registration and provides technical support for clinical applications.
ObjectiveThe relatively small fields of view of optical coherence tomography (OCT) and OCT angiography (OCTA) not only limit their clinical applications for disease diagnostics, but also lead to incorrect diagnoses. To achieve ultrawide-field OCT/OCTA imaging, two strategies are typically employed: one involves enhancing the optical design to expand the imaging field of view, which is complicated, expensive, and may introduce distortions that degrade the image quality; while the other involves performing multiple local scans with a flexible probe (such as a handheld probe), which can introduce motion artifacts. To obtain large-scale, high-quality OCT images, both flexible and stable scanning mechanisms besides high-precision image registration techniques are essential. Accordingly, in this study, a large-scale OCT imaging technique based on a 6-joint robotic arm is explored. First, the OCT probe is loaded and moved to multiple local regions for optical scanning. The resulting images are then precisely stitched using a dual-cross-correlation-based translation and rotation registration (DCCTRR) algorithm considering the coordinate information of the robotic arm. This research can serve as a valuable reference for improving the clinical applications of OCT, providing methods to enhance both the user experience and the overall effectiveness of OCT system techniques.MethodsA home-built spectral-domain OCT (SDOCT) system (Fig. 1) and a commercially available 6-joint robotic arm are adopted to test the proposed technique. The transformation matrix from the robot end effector to the OCT coordinate system is calculated using singular value decomposition (SVD). Consecutive local OCT scanning is performed using a home-developed C++ application, and the target pose is converted to a joint pose via an inverse kinematic calculation for robot pose control. To complete a large-scale scan of a chicken breast, 5×5 square grids covering ~8.2 mm×8.2 mm are set, and the overlap ratio can be flexibly adjusted for the registration algorithm mentioned above. Finally, 25 local OCT images are obtained and used as stitches to validate the performance of the proposed technique.Results and DiscussionsTo determine the coordinate transformation from the robotic arm end effector to the OCT coordinate system, the displacement of the steel ball center is measured during the three positional changes of the mechanical arm (Table 1). Regarding OCT image registration, the registration accuracy of 91.07% is achieved using the DCCTRR algorithm, significantly outperforming the kinematic matrix method with the accuracy of 77.20% (Fig. 4). Using the transformed information from the mechanical arm and the DCCTRR method, a large-scale frontal structural image is obtained from the registered 5×5 grid of local chicken breast OCT images (Fig. 6).ConclusionsIn this study, the use of a 6-joint robotic arm to load a high-resolution OCT system probe is explored with the aim of achieving large-scale, high-resolution imaging. Because the positioning accuracy of the robotic arm is lower than the OCT imaging resolution, post-image registration (using the DCCTRR algorithm) is required for high-precision image registration. Compared with manual operations, this approach can greatly improve the imaging field without introducing motion artifacts. In summary, robotic arms, image-registration algorithms, and flexible OCT probes are considered in this work to achieve large-scale high-resolution imaging. We believe that this research can serve as a valuable reference for improving the clinical applications of OCT, providing methods to enhance both the user experience and the overall effectiveness of OCT system techniques.
ObjectiveThe optical parameters of biological tissues can reflect their physiological state to a certain extent and provide an important reference basis for clinical diagnosis. Therefore, it is of great significance to measure the optical parameters of biological tissues. The commonly used methods for measuring the optical parameters of biological tissues have problems. Diffusion optical tomography has a deep imaging depth, but it relies on the depth learning algorithm of the simulated dataset, and its accuracy in practical applications is debatable. Optical coherence tomography, which has a high measurement accuracy, is only applicable to the measurement of optical parameters of shallow tissues. The direct measurement of scattering coefficients using a transmission model leads to a large error, and it cannot meet the requirements for measurement accuracy. Acousto-optic tomography (AOT) effectively combines the advantages of optical and acoustic technologies, and is expected to realize high-precision quantitative measurement of scattering coefficients of thick tissues. In this study, the feasibility of using acousto-optic signals to measure the scattering coefficients of tissues is confirmed by theory, finite element simulation, and experiment, and the advantages and disadvantages of the two types of measurement methods based on acousto-optic signals are compared.MethodsCombining the diffuse theory of light propagation in biological tissues with the intensity modulation mechanism of acousto-optic interaction, the relationship between acousto-optic signals and the scattering coefficient is obtained. The finite element software COMSOL Multiphysics is used to simulate the acousto-optic process in the tissue to verify the correctness of the theoretical analysis results. In the AOT experiment, the peak-to-peak value and relative intensity of the acousto-optic signals are obtained by fixing the incident intensity and changing the incident intensity, respectively. Combining the relationship between acousto-optic signals and the scattering coefficient, the quantitative measurement of the scattering coefficient of the simulated tissue fluid is realized.Results and DiscussionsIn the COMSOL Multiphysics simulation and AOT experiment, the peak-to-peak value of the acousto-optic signal shows a linear increasing relationship with the incident intensity (Fig. 5 and Fig. 10), and reveals an exponential decay trend with the scattering coefficient [Fig. 6(b) and Fig. 11(b)]. The relative intensity of the acousto-optic signal does not change with the change of the incident intensity (Fig. 5 and Fig. 10), and shows the same exponential decay relationship with the scattering coefficient [Fig. 6(a) and Fig. 11(a)]. The scattering coefficient of the medium is measured by the peak-to-peak value and relative intensity of the acousto-optic signal obtained by the simulation. The relative errors of the scattering coefficients obtained by both methods are within 0.5% (Fig. 7). The measurement accuracy of the former method is slightly better than that of the latter in the COMSOL Multiphysics simulation. In the AOT experiments, the maximum absolute error obtained using the relative intensity measurement method is 0.26 cm-1, the average absolute error is 0.10 cm-1, the maximum relative error is 3.88%, and the average relative error is 1.32% [Fig. 12(a)]. The maximum absolute error obtained using the peak-to-peak measurement method is 0.31 cm-1, the average absolute error is 0.12 cm-1, the maximum relative error is 3.34%, and the average relative error is 1.35% [Fig. 12(b)]. Under the same conditions, the measurement range of medium scattering coefficients using the relative intensities of acousto-optic signals is larger than that using the peak-to-peak values of acousto-optic signals [Fig. 13(a)].ConclusionsIn this study, the quantitative relationships between the peak-to-peak value and relative intensity of acousto-optic signals and the scattering coefficient of tissues are obtained. The peak-to-peak values of the acousto-optic signals show a linear incremental relationship with the incident intensity, but the relative intensity remains unchanged with the change in incident intensity. The relative intensity and peak-to-peak values of the acousto-optic signals show the same exponential decay trend with the increment of the scattering coefficient. The theoretical conclusions are verified through a COMSOL Multiphysics simulation and experiment. In the COMSOL Multiphysics simulation, the relative errors of the scattering coefficients based on the peak-to-peak values and relative intensities of the acousto-optic signals are both within 0.5%. In the AOT experiment, the maximum relative error of the scattering coefficient measured using the relative intensity of the acousto-optic signal is 3.88%, and the average relative error is 1.32%. The maximum relative error of the scattering coefficient measured using the peak-to-peak value of the acousto-optic signal is 3.34%, and the average relative error is 1.35%. It can be observed that the measurement accuracies of the two methods are comparable. In practice, the peak-to-peak value measurement method is fast, but the relative intensity measurement method can measure a larger range of the scattering coefficient. The above conclusions initially indicate the feasibility of high-precision quantitative measurement of scattering coefficients of biological tissues using acousto-optic signals. This is expected to provide a novel and non-invasive technical means for detecting biochemical attributes such as blood glucose, triglyceride, and total cholesterol concentrations in human blood tissues and can provide a certain reference for the clinical diagnosis of related diseases.
ObjectiveTumor organoids, as novel in vitro tumor models, hold significant value in tumor biology research and personalized drug sensitivity assessment. However, existing methods relying on manual seeding and destructive endpoint testing are limited by the lack of dynamic monitoring capabilities and the requirement for high sample homogeneity. This study aims to develop a non-destructive, dynamic analysis framework for tumor organoids based on 3D optical coherence tomography (OCT) and deep learning, enabling precise segmentation, morphological characterization, and growth analysis of organoids to assess drug responses efficiently.MethodsWe presented a label-free OCT-based framework that includes deep learning-driven segmentation, 3D morphometric quantification of individual organoids, and growth rate modeling of organoid clusters. To tackle 3D discontinuities in organoid segmentation, we introduced a novel parallel encoder architecture, ParaSAM2CNN, which integrates ResNet's deep feature extraction with SAM2's multiscale feature capture, enabling automated and precise segmentation (Dice coefficient: 0.8026). An adaptive surface roughness quantification algorithm was developed to enable longitudinal, high-throughput, multidimensional morphological characterization of organoids. Unsupervised clustering was applied to categorize organoid phenotypes, while principal component analysis (PCA) was employed to elucidate correlations among morphological parameters, growth dynamics, and drug response. A growth level model for organoid clusters was established and validated against traditional destructive ATP-based assays, showing high consistency (90.45%).Results and DiscussionsThe proposed framework demonstrates significant advantages in the non-destructive analysis of tumor organoids and drug response assessment. The ParaSAM2CNN model achieves superior segmentation performance compared to other state-of-the-art models, with improved precision and Jaccard index. The adaptive surface roughness algorithm provides detailed morphological characterization, capturing changes in organoid structure under drug treatment, such as the transition from cystic to solid phenotypes. The growth level model shows a high correlation with ATP test results, confirming its reliability in assessing organoid growth and drug sensitivity. This framework not only provides a non-invasive alternative to traditional endpoint testing but also offers a transformative potential for drug screening and personalized therapy optimization based on patient-derived tumor organoids.ConclusionsThis study presents a significant advancement in the analysis and application of tumor organoids for cancer research and treatment. By integrating OCT imaging with deep learning and machine learning techniques, we have developed a comprehensive and non-destructive evaluation framework that accurately assesses organoid growth and drug responses. This method has the potential to revolutionize traditional drug screening and sensitivity testing methods, providing a new technological platform for cancer research and personalized medicine. The high consistency with ATP testing highlights the model’s potential as a reliable and non-invasive tool for cancer treatment.
ObjectiveIn optical coherence elastography (OCE) based on an elastic wave, the phase velocity of the elastic wave is closely related to the Young modulus of tissues. The elastic properties of tissues can be derived by calculating the phase velocities of elastic waves at different frequencies to obtain the phase velocity dispersion curves. A high-frequency elastic wave plays a significant role in tissue elasticity characterization. On one hand, the spatial resolution of elastography is influenced by the elastic wave wavelength, where a higher frequency enables a higher spatial resolution. On the other hand, the wave velocity dispersion over an extended frequency range enables the extraction of detailed mechanical properties including the depth-dependent variation of elasticity and internal stress. However, the conventional two-dimensional Fourier transform (2D-FT) methods exhibit limitations in processing high-frequency elastic waves, because they are susceptible to noise interference, leading to the reduced accuracy in phase velocity estimation and consequently affecting the precise characterization of tissue mechanical properties like elasticity. In this work, we propose a phase velocity estimation method combining the generalized Stockwell transform with the Radon transform (GST-RT). The derived phase velocity dispersion curve is used to calculate the Young modulus. The generalized Stockwell transform is applied to perform a time-frequency analysis on the spatiotemporal displacement data. Subsequently, the Radon transform is applied to identify the elastic waves within the time-frequency-transformed spatial data at each frequency and calculate their phase velocities, thereby enabling the determining of the Young modulus. The feasibility of this method is validated through simulation data, and the robustness is evaluated in the presence of additive Gaussian white noise conditions. Additionally, the experimental analysis of agar phantom is conducted to assess the performance of the proposed method in practical applications.MethodsThe flowchart of the GST-RT method is shown in Fig. 1. First, the two-dimensional spatiotemporal displacement data are obtained from the optical coherence tomography (OCT) data with a phase-sensitive detection algorithm. Subsequently, the generalized Stockwell transform is used to map the spatiotemporal displacement data into a three-dimensional time-frequency-spatial domain. The Radon transform then converts the time-spatial data from the frequency domain to the angle-normal distance space. To validate the method, the GST-RT and 2D-FT methods are applied to elasticity simulation data under noise-free and noisy conditions (signal-to-noise ratios of 35 dB, 30 dB, 25 dB) for phase velocity estimation, and the percentage errors in phase velocity and Young modulus for both methods are calculated. Additionally, the agar phantom elasticity measurement experiments are conducted to validate the capability of the GST-RT method for evaluating the elastic properties of real samples. The Young modulus values calculated by both the GST-RT and 2D-FT methods are compared with those obtained from the mechanical compression tests.Results and DiscussionsThe simulation reveals that both methods achieve the phase velocity percentage errors below 1% and the Young modulus errors under 2% in noise-free conditions. The GST-RT demonstrates a robust performance even with added Gaussian white noises, exhibiting the phase velocity errors consistently within 1% and the Young modulus errors below 2%. In contrast, the 2D-FT method displays significantly higher errors, reaching up to 9.62% for phase velocity and 12.35% for Young modulus under noisy conditions, thereby confirming the superior robustness of GST-RT. The agar phantom experimental results demonstrate that the percentage errors in Young modulus calculated using the GST-RT method are consistently controlled within 2%, outperforming those of the 2D-FT method which exhibits a maximum error of 6.16%. These findings align with the simulation results, further validating the reliability and accuracy of the GST-RT method in analyzing experimental data.ConclusionsThis study proposes a phase velocity estimation method integrating the generalized Stockwell transform and the Radon transform. The generalized Stockwell transform captures high-frequency information while mitigating noise interference in calculations, while the Radon transform enables precise identification of elastic waves, thereby improving the accuracy of phase velocity estimation and enhancing the reliability of tissue elasticity characterization. Simulations validate the feasibility of the method. Comparative analyses under noise-free and noisy conditions demonstrate that the GST-RT method keeps Young modulus percentage errors below 2% in both conditions, highlighting its robust noise resistance. Agar phantom experiments confirm the method practicality. Compared to the conventional 2D-FT method, the GST-RT method demonstrates enhanced robustness in elastic simulation data contaminated with Gaussian white noises, while also extending the bandwidth of phase velocity analysis in experimental results. This approach provides a reliable quantitative and analytical method for tissue elasticity characterization.
ObjectiveThis study utilizes a dual-wavelength photoacoustic microscopic imaging system to effectively separate Evans blue dye within tissues. We establish a localized blood-brain barrier (BBB) disruption model in mice and conduct in vivo imaging investigations. Our results indicate that, even with an intact skull, the system can clearly present cerebral vascular structures and accurately identify the areas of BBB damage, demonstrating its significant advantages in the study of cerebrovascular diseases. With its high resolution and sensitivity, photoacoustic microscopy holds great promise for applications in the mechanistic research, early diagnosis, and treatment monitoring of brain diseases, particularly those affecting the brain vasculature and the blood-brain barrier, positioning it as a potential novel diagnostic tool.MethodsA dual-wavelength (532 nm/610 nm) photoacoustic microscope is established, and a localized BBB disruption model in mice is induced using a hot water stimulation method. Initially, the photoacoustic microscope is utilized to observe the BBB leakage process in the mouse ear. Subsequently, imaging is conducted on both experimental and control groups exhibiting BBB disruption, followed by quantitative analysis.Results and DiscussionsPhantom experiments validate the feasibility and accuracy of the spectral separation algorithm, as evidenced by the results shown in Fig. 3. The permeability detection experiment of ear blood vessels confirms the system capability to detect Evans blue extravasation from the blood vessels (Fig. 4). Tests on BBB integrity show that, even with an intact skull, the system can clearly image brain vascular structures with distinct regional boundaries, demonstrating its effectiveness in brain vascular imaging and assessing BBB integrity (Fig. 5).ConclusionsWe successfully develop a dual-wavelength photoacoustic microscopy imaging system based on 532 nm and 610 nm, achieving high-resolution imaging of mouse brain microvascular structures while preserving the skull. By incorporating spectral separation algorithms, this study quantifies the changes in BBB integrity. Photoacoustic imaging (PAI) shows great potential for researching brain diseases, particularly those affecting blood vessels and BBB, and is anticipated to become a vital tool in the fundamental research of brain disorders.
ObjectiveDislocation, distortion, and honeycomb artifacts, common problems in confocal endomicroscopic images, disturb the pixel arrangement of images and obscure potential information, thus hindering further analysis of the target object in an image. Many studies have focused on solving one of these individual problems, such as correcting image dislocation or distortion, or simply studying methods to remove honeycomb artifacts. The methods of dislocation correction and distortion correction mainly include hardware correction or post-correction of images using specific algorithms. Hardware methods increase the complexity of the system, whereas software algorithms are relatively time consuming. Methods to remove honeycomb artifacts include interpolation, filtering, image mosaicing, and deep learning; however, related research usually does not consider the impact of image dislocation and distortion. The applicability of some of these methods may be reduced when multiple problems exist simultaneously. A simple combination of multiple methods in a specific order is a feasible solution; however, it may lead to low image reconstruction efficiency. Therefore, developing efficient integrated solutions for mitigating the effects of dislocations, distortions, and honeycomb artifacts in confocal microscopy images is crucial. In this study, we proposed a confocal endomicroscopy image reconstruction method that efficiently combines dislocation correction, distortion correction, and honeycomb artifact removal to overcome multiple problems observed in confocal endomicroscopic images and recover potential information in images.MethodsThe proposed method consists of calibration and reconstruction stages. In the calibration stage, the proposed method obtained the dislocation of pixels, reference coordinates of the fiber core center, and the relative transmittance of the fiber core from a uniform fluorescence image. In addition, distortion correction and Delaunay triangulation were performed on the fiber center coordinates during the calibration stage, and the barycentric coordinates of each pixel in the images were calculated. Calibration information was used to assist in the reconstruction of subsequent images. In the reconstruction stage, the entire dislocation of the image was corrected according to the dislocation determined in the calibration stage, and the actual gray value of the fiber core center was obtained by searching for the maximum gray value. The gray value of the fiber core center was further corrected by dividing it by the relative transmittance to eliminate the influence of the difference in the transmission characteristics of each fiber core. Then, the corrected gray values were individually mapped to Delaunay triangular grids, and the honeycomb artifacts were removed by barycentric interpolation to obtain the final reconstructed image. We studied the influence of dislocation, distortion, and honeycomb artifacts on the reconstructed image through simulation and quantitatively evaluated the reconstruction effect of our method. In practical experiments, we captured pictures of USAF-1951 resolution targets and reconstructed the images to verify the effectiveness of our method. In addition, we captured the images of plant leaves and animal fat, and reconstructed them to evaluate the effect of our method on biological samples.Results and DiscussionsThe simulation results show that dislocation and core transmittance differences (Fig. 5) lead to abnormal gray values in the reconstructed image. Different regions in an image have different degrees of distortion, which cannot be corrected by horizontal image scaling. Compared to other methods, our method effectively eliminates abnormal gray values, corrects distortion, removes honeycomb artifacts, and obtains the highest peak signal-to-noise ratio (PSNR) and structural similarity (SSIM) (Table 1). The reconstruction results of the USAF-1951 resolution target in the experiments (Fig.8) show that the method presented in this study recovers the structural information of the resolution target. Compared to the simple combination of dislocation correction, distortion correction, and Gaussian filtering, the brightness and contrast of the reconstructed images obtained using our method are higher (see Fig.8 (e)), and the contrast-to-noise ratio (CNR) is improved by approximately 100% (Table 2). In the reconstructed image, the approximate structure contour of the second element of the eighth group can be recovered. However, the information in the reconstructed image cannot be identified by adopting a simple combination of dislocation correction, distortion correction, and Gaussian filtering. This result implies that our method can effectively protect and utilize the information contained in the original image during the reconstruction process, allowing the recovery of the potential structural features. The experimental results of the biological samples (Fig. 9) show that our method effectively reconstructs the structure of the biological samples, even though the original image contains dislocation, distortion, and honeycomb artifacts, allowing identification of the characteristic information of the biological samples in the image.ConclusionsWe presented a comprehensive method for confocal endomicroscopic image correction and reconstruction. This method efficiently combines dislocation correction, distortion correction, and honeycomb artifacts removal, and avoids repeated dislocation evaluation and distortion correction through one-time calibration. The experimental results show that the proposed method can effectively eliminate image distortion and honeycomb artifacts introduced by fiber bundles, thus improving the image quality and recovering the structural details of objects in images. Compared to Gaussian filtering combined with distortion correction, this method can utilize the information of the original image more effectively, and the reconstructed image has a higher contrast and less noise.
ObjectiveThe accurate diagnosis of ophthalmic diseases largely depends on the comprehensive analysis of both structural and functional retinal features. Multimodal imaging techniques, such as multi-channel color fundus (MC) photography and fundus fluorescein angiography (FFA), provide complementary information. MC offers high-resolution anatomical details, whereas FFA highlights functional abnormalities like vascular leakage and non-perfusion areas (NPAs). However, existing deep-learning-based fusion methods have critical limitations. First, they fail to fully exploit the spectral properties of retinal tissues, which are essential for distinguishing subtle pathological features. Second, conventional approaches rely on color space conversion (e.g., YCbCr) to generate single-channel fusion results, leading to irreversible color distortion and the loss of multi-channel depth information. Third, the limited receptive fields of convolutional neural networks (CNNs) hinder the effective integration of multi-scale features, particularly for small but clinically significant structures such as capillaries (typically <5 μm in diameter) and microaneurysms. These challenges necessitate a novel fusion framework that preserves both high-frequency vascular details and low-frequency background textures while maintaining human visual consistency. This study addresses these gaps by proposing a diffusion-model-based fusion framework tailored for ophthalmic imaging, with the aim of enhancing diagnostic accuracy through high-fidelity multimodal integration.MethodsThe proposed framework integrates a denoising diffusion probabilistic model (DDPM) with frequency-adaptive dilated convolution (FA Conv) to achieve anatomically precise and perceptually consistent fusion of MC and FFA images. First, a four-channel input tensor is constructed by concatenating the three-channel MC and one-channel FFA images, bypassing color space conversion to prevent information loss. The DDPM learns the joint distribution of MC and FFA through a forward-reverse diffusion process (Fig. 2). In the forward phase, Gaussian noise is incrementally added to the input over 1000 timesteps, mapping the original data to a latent space. The reverse phase employs a U-Net-based denoising network with residual blocks and positional encoding to predict the noise at each timestep, effectively extracting the multi-scale diffusion features (Fig. 3). To address the tradeoff between bandwidth and receptive field, the framework incorporates Fourier decomposition and dynamic dilated convolution. The high-resolution features from the diffusion decoder are decomposed into four frequency bands using a discrete Fourier transform. Adaptive dilation rates are assigned per pixel based on high-frequency energy, enabling larger receptive fields for low-frequency regions (e.g., retinal background) and finer sampling for high-frequency structures (e.g., capillaries). The fusion weights are dynamically adjusted using a learnable frequency selection map, ensuring optimal integration of cross-modal features (Fig. 6). Finally, a multi-scale loss function combines gradient preservation, intensity alignment, and VGG-19 perceptual similarity to maintain color fidelity and structural coherence. The two-stage training strategy first optimizes the DDPM for noise prediction (Algorithm 1, Fig. 4) and then fine-tunes the fusion module with perceptual constraints (Algorithm 2, Fig. 7).Results and DiscussionsThe proposed method is evaluated on a dataset encompassing 75 eyes with 1500 image pairs. Qualitative comparisons (Figs. 8?11) highlight the superior performance of our method in preserving the intricate textures of the MC images and the pathological features of the FFA images. For instance, in cases of central serous chorioretinopathy, the fused images distinctly exhibit microaneurysms in the MC images alongside vascular leakage in the FFA images, offering a comprehensive depiction of the disease. Similarly, in retinal artery occlusion scenarios, the integrated images highlight obstructed blood vessels and changes in the surrounding tissue, aiding in precise diagnosis. Quantitative evaluations (Table 1) substantiate the advantages of our approach. Our method surpasses the leading fundus fusion techniques in multiple metrics, including entropy (EN, 6.669), standard deviation (SD, 67.497), correlation coefficient (CC, 0.899), multi-scale structural similarity index measure (MS-SSIM, 1.386), learned perceptual image patch similarity (LPIPS, 0.347), and Delta E 2000 (5.009). These results suggest that our approach produces fused images with richer informational content, heightened contrast, superior structural preservation, and more precise color representation. Furthermore, ablation studies (Table 2) are executed to evaluate the efficacy of the diffusion model and frequency-adaptive convolution. The findings reveal that omitting the diffusion model significantly compromises color accuracy, whereas eliminating the frequency-adaptive convolution reduces structural preservation. This highlights the pivotal role of both components in ensuring high-fidelity fusion of multimodal fundus images.ConclusionsThis paper introduces the first diffusion-based fundus image fusion framework capable of achieving high-fidelity multimodal fusion of MC and FFA modalities. By leveraging the multi-scale feature extraction capabilities of DDPM alongside the dynamic balancing mechanisms of frequency-adaptive convolution, the proposed method successfully integrates these two modalities while preserving both anatomical structures and lesion-specific information. This results in fused images that maintain visual consistency for human observers, making them more suitable for clinical diagnosis. These findings suggest that our proposed method has the potential to enhance the diagnostic accuracy and reliability in ophthalmology, particularly for diseases requiring multimodal analysis. Future research could explore the integration of additional retinal imaging techniques, such as optical coherence tomography angiography (OCTA) or three-dimensional (3D) modalities, to provide a more comprehensive visualization of the retina. Moreover, the proposed framework can be adapted to other medical imaging applications where multimodal image fusion is crucial for precise diagnosis and treatment planning.
ObjectiveBurns are a common type of skin injury. Diagnosing the degree of the burn is very important for proper treatment. Optical coherence tomography is a non-invasive, non-destructive, and high-resolution optical detection technology. Polarization-sensitive optical coherence tomography (PS-OCT) provides a comparison of birefringence information compared to the conventional structural OCT modality. It can be used for the high-resolution, high-contrast, real-time three-dimensional imaging of damaged skin. In this work, a simple, compact, flexible, and efficient PS-OCT system is developed based on single-mode fiber optics with a circularly polarized single-input state as the swept source. The high-performance swept source enables a high imaging speed and long coherence length for the OCT imaging. The PS-OCT system is based on single-mode fiber optics and features low polarization crosstalk, low polarization mode dispersion, and a compact size. A multiple-parameter analysis shows that the PS-OCT system has the potential to provide accurate clinical assessments of skin burns.MethodsWe construct a swept-source PS-OCT system with single-mode fiber optics. By tuning the polarization controllers step by step, a single circular polarization input in the sample arm and OCT signal detection with orthogonal polarization channels are achieved. Using straightforward data processing algorithms, the PS-OCT system has the capability to acquire various parameters, including the structural intensity, degree of polarization uniformity (DOPU), cumulative phase retardation (CPR), and Stokes state. Given the anatomical and physiological resemblance between pig skin and human skin, ex vivo pig skin is selected as the imaging subject for the skin burn model in this study. To simulate the burns, eight groups of pig skin samples are subjected to a circular thermal injury with a diameter of 10 mm using a temperature-controlled wound burning device at 90 °C for a duration of 30 s. We compare the multi-parameter PS-OCT images of the normal and burned pig skin samples. According to the image histogram, the Bhattacharyya distance is calculated to demonstrate the capability of the PS-OCT system for skin burn evaluation.Results and DiscussionsIn the structural OCT images, the difference between the normal and burned pig skin samples is not obvious (Fig.3). As shown in the cross-sectional structural OCT images, the total scattering intensity has similar values in the regions of the normal and burned skin samples. In the en-face structural images, the boundary of the burned skin region is clear, and the pattern of the skin texture is different. Compared to the structural images, the polarized images show obvious differences between the normal and burned pig skin samples in terms of the Stokes state, DOPU, and cumulative phase retardation (Fig.4). In the region of the burned skin, the color of the Stokes state image becomes relatively uniform, the value of the DOPU image is relatively large, and the CPR value is relatively low. The en-face images demonstrate that the structural intensity values of the normal and burned pig-skin regions are very similar, whereas the DOPU, CPR, and Stokes values have relatively large differences. The histograms of these en-face images further verify that polarized images are more useful in distinguishing normal and burned skin (Fig.5 and Fig.6). Using to the histograms, we calculate the Bhattacharyya distance to quantify the difference in the images between normal and burned pig skin (Fig.7). If the images are very similar, the Bhattacharyya distance is close to 0. If the images are very different, the Bhattacharyya distance is close to 1. For the 8 groups of skin burn experiments, the average Bhattacharyya distance of the structural images is 0.184, while the values for the DOPU images, CPR images, and Stokes images are 0.917, 0.744, and 0.839, respectively. A quantification analysis demonstrates that the difference between normal and burned skin in the traditional OCT structural images is small, while the polarized images show a significant difference in the burned skin. The PS-OCT system used in this study adopts a design based on single-mode fiber optics. However, the birefringence characteristics of single-mode fiber optics are easily affected by environmental factors such as bending stress and temperature changes. Therefore, once we have completed the steps to calibrate the polarization state of the PS-OCT system, the optical fibers in the system must not be touched. In an actual work environment, the polarization state of the imaging system can be maintained for several days without significant changes, thus meeting the needs of most clinical and life science applications. In the future, we will further optimize the optical design of the PS-OCT system to improve its polarization stability. In addition, the PS-OCT imaging system shows that a polarized image of pig skin tissue exhibits an obvious change after being burned. The mechanism of the change in the polarization state mainly comes from the irreversible denaturation of the collagen and elastic fibers in the skin tissue after heating. However, in this study, only a small amount of ex vivo pig skin is used as the model for a skin burn. The number and type of experimental samples are insufficient. In addition, the ex vivo skin samples have lost their biological tissue activity characteristics. In the future, we will increase the number and type of skin burn models. Furthermore, we also need to investigate living animal samples and skin burn patients to promote the application of PS-OCT imaging in the diagnosis and treatment of skin burns.ConclusionsA flexible and efficient PS-OCT system based on single-mode fiber optics and a single-state input is built to image ex vivo pig skin for skin-burn investigations. The system can provide structural images and three polarized images (DOPU, CPR, and Stokes state) of skin tissue. We compare images of normal and burned skin, and perform histogram statistical analysis to illustrate the distribution of these parameters. Moreover, we calculate the Bhattacharyya distance as a histogram similarity coefficient to further quantify the imaging performance. The results show that there are significant birefringence changes in the burned skin tissue compared to the normal skin tissue, which are mainly due to the denaturation of the collagen and elastic fibers after heating. The changes in burned skin can be clearly observed using the polarization parameters (DOPU, CPR, and Stokes state). These polarized OCT images exhibit enhanced contrast and more pronounced distinctions for burned skin compared to conventional structural OCT images. This research demonstrates the promising potential of PS-OCT technology for skin-burn diagnosis.
ObjectiveQuality testing of tomatoes is critical in many aspects of their growth, storage, and transportation. Sugar content (Brix) is a necessary criterion for evaluating whether a fruit is tasty. The timely and accurate mastery of quality parameters, such as the Brix distribution during the ripening process, is crucial for the scientific and efficient cultivation of tomatoes. Among the widely used detection methods, the Brix meter detection, which is based on the principle of light refraction, is a mean value measurement method based on the fruit juice, which cannot satisfactorily assess the fine distribution of different parts of a sample. The hyperspectral imaging technology leads to a significant amount of redundancy in the amount of data and is easily affected by the depth of light penetration and the water content of the sample, which is insufficient to detect and characterize the full range of details of the sample. Mass spectrometry is complex and cumbersome for sample preparation and does not support rapid detection. This study proposes a method for detecting and characterizing the sugar content of tomato fruits from the perspective of cellular phase information based on phase imaging technology.MethodsThe physiological properties of the samples were analyzed and characterized in this study from a cellular perspective, starting from the microscopic material basis of plant growth. Based on the characteristics of the quantitative phase microscopy, which uses the inherent contrast source of different refractive indices between different components in the cell, we performed phase imaging experiments on tomato pulp cells and extracted the cellular phase parameters, called "phase envelope volume" and "phase peak." The correlation between the phase parameters and Brix was analyzed by comparing them with the measurement results of the Brix meter. Based on the experimental data, a data cube of the two-phase parameters and Brix was constituted, and positive correlations between the phase parameters and Brix were obtained. This provides a basis for characterizing local Brix using phase parameters.Results and DiscussionsThe proposed detection and characterization method can be applied to any part of a tomato plant and requires only a single frame of the phase map at any incidence angle. The extraction of phase parameters eliminates the tedious operation of decoupling the physical thickness and refractive index of the cells, and the entire analysis process only takes approximately 0.5 s. Based on the sensitivity of the phase information to the internal chemical composition of the cell, the phase parameter characterization method can also be applied to detect the physiological state of other fruits and vegetables in addition to sugar (Fig. 10). This study provides a reference for the refinement and precision in detecting the quality of agricultural products.ConclusionsThis paper focuses on the demand for rapid and accurate quality inspection of fruits and vegetables in modern agriculture. A detection method based on phase imaging is proposed. The inspection of tomato fruit sugar is used as an example to explain the related principles and procedures. In this method, only a single frame of the phase map from any angle of incidence is required. Two phase parameters, that is, "phase envelope volume" and "phase peak," are extracted from the phase map to quantitatively investigate the sugar content characteristic of a cell. The experimental results of the comparison with the detection based on the Brix meter indicate that the sugar content and the above phase parameters show a significant positive correlation. This provides a basis for characterizing the sugar content using the phase parameters. The local sugar content distribution of a tomato fruit was detected experimentally, and the results show good consistency with those of hyperspectral detection. The feasibility and effectiveness of this method have been demonstrated to a certain extent. This phase detection and characterization method requires only one frame of the phase image, and the related analysis process eliminates the cumbersome operation of decoupling the physical thickness and refractive index of the cell. This means that the hardware and time costs can be reduced. Based on the sensitivity of the phase information to the internal chemical composition of the cell, the phase parameter characterization method can also be applied to detect the physiological state of other fruits and vegetables, which may be used as a workable solution for the rapid and accurate detection of agricultural product quality.
ObjectiveCervical cancer is one of the most common malignant tumors and poses a serious threat to human health. However, because the onset of cervical cancer is gradual, early and effective screening is crucial. Traditional screening methods rely on manual examinations by pathologists, a process that is time-consuming, labor-intensive, error-prone, and often lacks an adequate number of pathologists for cervical cytology screening, making it challenging to meet the current demands for cervical cancer screening. In recent years, several deep-learning-based methods have been developed for screening abnormal cervical cells. However, because abnormal cervical cells develop from normal cells, they exhibit morphological similarities, making differentiation challenging. Pathologists typically need to reference normal cells in images to accurately distinguish them from abnormal cells. These factors limit the accuracy of abnormal cervical cell screening. This study proposes a Transformer-based approach for abnormal cervical cell screening that leverages the powerful global feature extraction and long-range dependency capabilities of Transformer. This method effectively enhances the detection accuracy of abnormal cervical cells, improving screening efficiency and alleviating the burden on medical professionals.MethodsThis study introduces a novel Transformer-based method for abnormal cervical cell detection that leverages the powerful global information extraction capabilities of Transformer to mimic the screening process of pathologists. The proposed method incorporates two innovative structures. The first is an improved Transformer encoder, which consists of multiple blocks stacked together. Each block comprises two parts: a multi-head self-attention layer and a feedforward neural network layer. The multi-head self-attention layer captures the correlation of the input data at different levels and scales, enabling the model to better understand the structure of the input sequence. The feedforward neural network layer includes multiple fully connected layers and activation functions and introduces nonlinear transformations to help the model adapt to complex data distributions. We also introduce Depthwise (DW)convolution and Dropout layers to the encoder. DW convolution layer performs convolution operations with separate kernels for each input channel, capturing features within the channels without introducing inter-channel dependencies. Dropout layer reduces the tendency of neural networks to overfit the training data, thereby enhancing the generalization of the model to unseen data. Additionally, we design a dynamic intersection-over-union (IOU) threshold method that adaptively adjusts the IOU threshold. In the initial stages of training, the model can obtain as many effective detections as possible, whereas in later stages, it can filter out most false positive predictions, thereby improving the detection accuracy of the model. Using the proposed method, the model can obtain precise information regarding the location of abnormal cells.Results and DiscussionsTo validate the effectiveness of our proposed method, we compare it with common general-purpose object detection methods. The average accuracy (AP) and AP50 of our method are 26.1% and 46.8%, respectively, surpassing those of all general object detection models (Table 1). In particular, our method outperforms other comparative models by a significant margin in AP metrics, demonstrating that our model not only detects normal-sized targets but can also detect extremely small targets. Additionally, in a comparison with attFPN, a network specifically designed for abnormal cervical cell detection, our method surpasses attFPN in terms of AP by 1.1% (Table 2). Visual inspection of the detection results reveals that our method more accurately identifies target regions with lower false-positive and false-negative rates (Fig.5). Ablation experiments indicate that adopting the improved Transformer encoder method increases AP and AP50 by 1.8% and 2.3%, respectively, compared with the original model. The use of dynamic IOU thresholds results in a 0.6% increase in AP and a 0.9% increase in AP50 compared with the original model (Table 4). Furthermore, a comparison between the dynamic and fixed IOU thresholds in terms of loss and AP during the training process shows that the model with dynamic IOU thresholds experiences a faster loss reduction and achieves a higher AP in the later stages of training (Fig.6).ConclusionsThis study introduces an automatic identification method for abnormal cervical cells utilizing Transformer as the backbone. We further propose an enhanced Transformer encoder structure and a dynamically adjustable IOU threshold. Various comparative experiments on datasets demonstrate that the proposed method outperforms existing approaches in terms of accuracy and other metrics, thereby achieving precise identification of abnormal cervical cells. Through ablation experiments, it is proven that both proposed modules enhance the accuracy of the model in identifying abnormal cervical cells. Overall, the proposed method significantly improves the efficiency of medical image screening, saving medical time and resources, facilitating timely detection of cancerous lesions, and presenting considerable clinical and practical value. Future research may focus on the application of semi-supervised and unsupervised learning in the field of medical imaging to enhance image utilization, improve model detection performance, and better meet clinical requirements.
ObjectiveThe accurate classification of white blood cells (WBCs) is crucial in the examination of blood and the diagnosis and treatment of clinical conditions. Manual examination under a bright-field microscope, the gold standard for blood cell analysis, is time-consuming and inspector-dependent. Currently, blood cell analyzers based on the impedance method or flow cytometry are extensively employed. However, some false positives may occur because of the structural variability of WBCs, which requires a manual microscopic review. In addition, these instruments are expensive. Deep learning, which can reduce the technical requirements of inspectors, is widely used for WBC classification. However, this analysis continues to rely on the morphology and color characteristics of the stained cells. To achieve high accuracy in the classification of WBCs, the process usually requires image acquisition and processing under a 100× objective lens, which can be time-consuming and data-intensive. Quantitative-phase imaging (QPI) is an effective method for studying cell morphology and biochemistry. However, identifying WBCs solely based on their phase characteristics is challenging, particularly when these phase characteristics are not prominent. Research on stained cells using QPI has shown that the inclusion of phase information, alongside bright-field pictures, might provide useful insights for WBC classification. In this study, the phase distributions of five different types of WBCs were quantitatively analyzed, and the substructure phase information was effectively divided using a co-localization imaging system based on digital holographic microscopy (DHM) and bright-field microscopy. A series of feature parameters were extracted to assist with the WBC classification. The accuracies of the classification of the three types of granulocytes based on the extracted phase feature parameters were 94%. Additionally, atypical lymphocytes were studied, and a recognition accuracy of 84.5% was achieved. The proposed method utilizes routine blood smear samples stained for clinical microscopy, making it easy to integrate into a commercial microscopic system and providing a wide range of practical applications.MethodsA benchtop co-localization imaging system was used to obtain bright-field images and quantitative phase images of WBCs from peripheral blood smears of healthy individuals. Quantitative phase images of the WBCs were reconstructed from off-axis holograms obtained from DHM. To segment the phase information, WBCs were first extracted and divided into two parts, the nucleus and the cytoplasm, based on bright-field images. Then, the position information of the nucleus and cytoplasm of the WBCs in the bright-field images was transposed onto the corresponding phase images. Finally, the quantitative phase distributions of WBCs and their corresponding nuclei and cytoplasm were successfully acquired. A substantial number of WBC samples consisting of 100 neutrophils, eosinophils, basophils, monocytes, large lymphocytes, and small lymphocytes were selected for co-localization imaging and statistical analysis. Various feature parameters were extracted to quantitatively describe and analyze the morphological and substructural features of the different WBCs.Results and DiscussionsThe feature parameters of the five types of WBCs were subjected to analysis and comparison, revealing distinct phase characteristics for each type. Neutrophils had a substantially higher nuclear phase value than the cytoplasmic phase value [Fig. 4(a)], whereas eosinophils had comparable nuclear and cytoplasmic phase values (Fig. 4). The cytoplasmic phase values in basophils fluctuated substantially [Fig. 5(c)], and monocytes showed a smaller phase difference between the nucleus and cytoplasm than lymphocytes [Fig. 4(b)]. Using the extracted feature parameters, three types of granulocytes were successfully classified with 94% accuracy. The efficiency of classifying phase features was evaluated by analyzing a total of 1200 neutrophils and eosinophils. This analysis was conducted using a phase feature method based on a 40× co-localization microscope, deep learning classification based on a 40× brightfield microscope, and a commercial system called Morphogo with a 100× microscope. The results showed that the phase feature accurately identified easily confused cells in deep learning classification or the Morphogo system (Fig. 7). Furthermore, an examination of atypical cells was conducted, revealing that the use of phase characteristics resulted in a classification accuracy of 84.5%. These results demonstrate that the phase feature parameters are effective in aiding WBC classification.ConclusionsThis study proposes a method for classifying WBCs using QPI. The approach involved analyzing different types of WBCs using a co-localization imaging system that combines DHM and bright-field microscopy. The position and structural information of WBCs were obtained from bright-field images, and the phase information of WBCs and their nuclei and cytoplasm were extracted accordingly. Statistical analysis was then used to extract feature parameters that effectively aided in the classification of WBCs. This method achieved an accuracy rate of 94% for classifying the three types of granulocytes based on the substructure phase characteristic parameters. Further analysis showed an accuracy rate of 84.5% for identifying atypical lymphocytes, which are often misinterpreted during microscopic examinations. Compared with using only phase information to classify WBCs, the proposed method incorporates high contrast between the nucleus and cytoplasm in bright-field images to effectively compare the characteristics of different WBC substructures, leading to an improved classification scope and accuracy. In addition, compared to conventional microscopic classification, the proposed method provides additional phase information that can assist in WBC classification. This method is easy to integrate with microscope and does not require the special treatment of conventionally stained blood smear samples. It is expected to be widely used for the leukocyte classification and diagnosis and treatment of various blood diseases.
ObjectiveThe nuclear pore complex (NPC) is an intricate structure comprising multiple distinct nuclear pore proteins known as nucleoporins (Nups). It plays a crucial role in the transformation of matter and information between the nucleus and cytoplasm. With a total molecular weight of 110‒125 MDa, the NPC is hailed as the "holy grail" of structural biology. Scientists have used such techniques as electron microscopy, atomic force microscopy, and cryoelectron microscopy to collectively reveal the composition, assembly, and ultrastructure of the NPC, providing a solid structural foundation for further exploration of its functions. The diameter of the NPC is approximately 130 nm. Therefore, single-molecule localization microscopy (SMLM) with an imaging resolution of 20 nm is an ideal tool for studying the ultrastructure of NPC. However, during long-term imaging, data loss may occur because of sparse blinking, and the dynamic activities of life also lead to heterogeneity in imaging results, posing challenges for data analysis. To address these issues, corresponding image reconstruction methods must be developed. Clustering algorithms are powerful tools for quantitative extraction, classification, and analysis of SMLM data. The unique clustered distribution structure of the NPC makes clustering methods highly suitable for structural analysis of the NPC. Therefore, to compensate for the limitations of SMLM data and obtain more detailed structural information about the NPC, a processing procedure for SMLM images of the NPC was developed in this study based on clustering algorithms. It involves screening out NPC structures with a more uniform morphology, followed by subjecting these structures to high-throughput statistical analysis and reconstruction.MethodsAfter PFA fixation, permeabilization with a blocking buffer, and labeling with antibodies (Nup133 and Nup98), U2OS cells were imaged by a self-built SMLM imaging system. A total of 50000 frames were captured after appropriate fields of view were selected. Through localization and drift correction processes, corresponding SMLM images were obtained. After the regions of interest were selected, the coordinate data with high localization accuracy were preserved for further analysis. First, a first round of density-based spatial clustering of applications with noise clustering (DBSCAN) analysis was used to remove background noise, identify individual NPCs, and determine the centroids of the NPCs (Fig. 3). To achieve a more accurate delineation of each Nup within every NPC in the case of retaining all signal points, a combination of the DBSCAN algorithm and hierarchical clustering was employed in the second round of delineation. In the second round of DBSCAN analysis, the algorithm was applied to identify the number of individual Nups within each NPC, and the data were further input into a hierarchical clustering algorithm for refinement of Nup localization. Subsequently, NPCs containing four to eight Nups were retained, and a second screening based on shape factors was performed to preserve NPCs with more uniform morphologies. Finally, the centroids of all remaining NPCs were aligned to obtain the complete distribution of labeled Nups in the NPCs. Using the least-squares method with NPC centroids as the center, a reconstruction of the Nup distribution with octagonal symmetry was achieved (Fig. 4). The reconstructed structure can be used to analyze the spatial characteristics of the Nup.Results and DiscussionsNup133, as a characteristic "Y"-scaffold-shaped component protein, has received extensive attention in recent research. Through statistical analysis of multiple datasets, the first round of the DBSCAN algorithm identified 10329 NPCs (Fig. 5). Among them, 3076 NPCs containing four to eight Nup133 were present, accounting for approximately 30% of the total. By selecting based on shape factors, a final set of 558 NPCs with relatively regular shape was obtained, accounting for approximately 5% of the total (Table 1). The retained NPCs were aligned by their centroids, resulting in an overlapped NPC image. Gaussian fitting was applied to calculate the radii of all Nup133, with the peak corresponding to a horizontal coordinate of (58.4±0.1) nm. This value is very close to the Nup133 radius of (59.4±0.2) nm calculated using the particle averaging method with antibody labeling. This further demonstrates the high-precision performance of the screening and reconstruction methods used in this study. In addition, the same analysis process was applied to analyze NPCs labeled with Nup98. Compared with that of Nup133, the distribution of Nup98 located in the inner ring of the NPC is more condensed (Fig. 6). A total of 10668 NPCs were analyzed, and 1126 NPCs were ultimately retained, accounting for approximately 10% of the total (Table 1). Similarly, the remaining NPCs labeled with Nup98 were aligned by the centroids, and Gaussian fitting was applied to the overlapped Nup98, resulting in a peak corresponding to a horizontal coordinate of (39.7±0.2) nm (Fig. 6). Compared with that of Nup133, the radius of Nup98 is smaller by 18.7 nm, indicating that Nup98 is closer to the center position of the NPC than Nup133. Finally, the eightfold symmetric structure of Nup133 and Nup98 was successfully reconstructed using the rotation alignment method, which is consistent with the acknowledged model.ConclusionsThe present study proposes a processing workflow based on clustering methods for screening and reconstruction of SMLM images of the NPC. The workflow has three main parts: classification, screening, and reconstruction. By performing two rounds of clustering to identify the NPC and Nup components, NPCs with a uniform shape containing four to eight Nups are selected and subjected to reconstruction analysis. The NPC with an eightfold symmetric structure is successfully reconstructed using the proposed workflow. Experimental results on Nup133 and Nup98 show that the radius of Nup133 is (58.4±0.1) nm, which closely aligns with the radius determined by the particle averaging method. The radius of Nup98 is (39.7±0.2) nm, indicating that Nup98 is situated in closer proximity to the central region of the nuclear pore. The proposed method reproduces the eightfold symmetric structure of the NPC, providing accurate localization information and aiding in a deeper understanding of the composition of this important structure. This clustering-based reconstruction method can also be extended to other nuclear pore-like structures, such as centrioles and basal bodies, or other structures with isotropic symmetric features, offering important strategies and methods for deciphering complex biological structures.
SignificanceIn recent years, the rapid development of bioimaging technology has provided powerful tools for life science research. Among them, fluorescence imaging, as an important imaging technique, enables real-time and non-invasive visualization of physiological activities in biological systems. Since biological tissues have lower absorption and scattering of photons in the near-infrared region Ⅱ (NIR-Ⅱ), combined with the weaker autofluorescence of tissues in this region, the signal-to-background ratio is greatly improved. Therefore, NIR-Ⅱ fluorescence imaging can achieve deeper and higher-resolution biological imaging, and is expected to be widely used as an ideal precision imaging technique in basic research and clinical practice in the future.NIR-Ⅱ fluorescence probes can be mainly categorized into inorganic and organic probes. Organic probes have advantages such as strong near-infrared absorbance, good biocompatibility, and easy metabolism, making them the preferred choice for in vivo imaging. Currently, there are two major classes of organic probes used in NIR?Ⅱ fluorescence imaging. One class is dyes with a donor-acceptor-donor structure, and the other class is cyanine dyes connected by conjugated polymethyl chains with a certain length of carbon atoms. Compared with donor-acceptor-donor dyes, the synthesis process of NIR-Ⅱ cyanine dyes is relatively simple, and they have higher brightness, thus possessing significant advantages in NIR-Ⅱ imaging. Cyanine dyes, as a highly valuable class of molecular probes, exhibit excellent fluorescence characteristics in the NIR-Ⅱ region, so that they have attracted extensive research interests and continue to develop in the field of disease diagnosis and treatment. Due to the high tissue penetration depth and low interference from biological background signals, NIR‐Ⅱ cyanine dyes can overcome the drawbacks of traditional fluorescence probes and be applied in the diagnosis of diseases.Cancer is one of the leading causes of death of the global population, characterized by high mortality and recurrence rates. NIR-Ⅱ cyanine dyes can be used for tumor detection and visualization in the NIR-Ⅱ region. Their high sensitivity and high-resolution imaging make them important tools for early tumor diagnosis. Additionally, cyanine dyes also have significant advantages in real-time dynamic display of tumor boundaries, providing critical information for tumor resection surgery. Inflammation is a protective response to stimuli. However, if inflammation persists without timely diagnosis and effective control, the detrimental effects will outweigh its biological benefits. NIR-Ⅱ cyanine dyes also have important value in the application of inflammatory diseases. On the one hand, they can precisely locate the inflammatory area, and on the other hand, by monitoring the distribution and concentration changes of the dye in the body, the activity of inflammation can be evaluated, providing guidance for treatment. NIR-Ⅱ cyanine dyes can also effectively differentiate and locate the sites of injury, visualize injuries, and help assess the extent of tissue injuries. Furthermore, the combination of NIR-Ⅱ cyanine dyes with drugs enables targeted drug delivery to tumors, inflammatory areas, or injured sites. By monitoring the distribution of the drug-dye complexes in the body, the therapeutic effect can be assessed in real time. These advanced applications demonstrate the tremendous potential of NIR-Ⅱ cyanine dyes in the field of modern medicine and their broad application prospects in diseases.ProgressThe latest advancements in the applications of NIR‐Ⅱ cyanine dyes in various diseases are summarized. First, the structural characteristics, classifications, and applications of cyanine dyes are introduced. Then, the utilization of NIR-Ⅱ cyanine dyes in various tumors such as brain tumors, breast cancer, pancreatic cancer, liver cancer, bladder cancer, colorectal cancer, gastric cancer and other tumors is comprehensively reviewed, referencing prior researches. Tian’s research group from University of Chinese Academy of Sciences and Gambhir et al. from Stanford University have developed an integrated visible and NIR-Ⅰ/Ⅱ multispectral imaging instrument to perform the first human liver tumor surgery. They have taken relatively pioneering studies on treatment of tumors. Additionally, the research group from Fudan University, led by Zhang, has developed a tumor-microenvironment-responsive lanthanide-cyanine fluorescence resonance energy transfer (FRET) sensor for NIR-Ⅱ luminescence-lifetime in situ imaging of hepatocellular carcinoma [Fig. 5(a)]. The applications of NIR-Ⅱ cyanine dyes in various inflammatory diseases like acute vascular inflammation, rheumatoid arthritis, gastritis, and in injuries related to liver, kidney, and biliary tract are further discussed. Liu et al. collaborated on cyanine-doped lanthanide metal-organic frameworks for NIR-Ⅱ bioimaging [Fig. 6(a)]. It is noted that the application research of NIR-Ⅱ cyanine dyes is still limited and not comprehensive. Finally, the challenges and research trends in this field are discussed.Conclusions and ProspectsNIR?Ⅱ cyanine dyes have tremendous potential in the imaging and treatment of tumors, inflammatory diseases, and injuries. In summary, further in-depth exploration is needed for the development of NIR-Ⅱ cyanine dyes to promote their wider clinical applications, and bring new breakthroughs and developments to the fields of medical imaging and clinical diagnosis.
SignificanceBecause of the wave characteristics of light, conventional fluorescence microscopy is typically restricted by the diffraction limit, which is approximately 200 nm laterally and 500 nm axially. Super-resolution microscopy has overcome this barrier and improved the imaging resolution to a few nanometers, which enables the observation of biological structures at a nanoscale and revolutionizes the development of life sciences. Super-resolution microscopy can be classified into three types. The first type is scanning imaging based on point spread function (PSF) decoration, whose representative technique is stimulated emission depletion (STED). The second is wide-field imaging based on spectrum spread, whose representative technique is structured illumination microscopy (SIM). The third is single-molecule localization microscopy (SMLM), also known as photoactivated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM). In super-resolution fluorescence microscopy, both instrumentation and sample-induced aberrations decrease the spatial resolution and degrade the imaging quality. Therefore, the adaptive optics (AO) technique is applied, which detects aberrations using direct or indirect methods, and performs compensation through wavefront correction elements to capture high-quality super-resolution images (Fig.1). This review introduces the origin and working principle of AO, summarizes its application in super-resolution fluorescence microscopy, and highlights its future development prospects.ProgressIn STED microscopy [see Fig.2(a)], aberrations in the excitation, depletion, and emission paths influence the image quality simultaneously, particularly in the depletion path, which need to be corrected with AO. In 2012, Gould et al. proposed the first implementation of AO in STED microscopy, which used modal sensing with the sharpness metric to examine the aberrations, and performed corrections with two spatial light modulators (SLMs) in both excitation and depletion paths [see Fig.2(b)]. They imaged fluorescence beads at a depth of 25 μm above the retina sections of a zebrafish, with an axial resolution of 250 nm. In 2014, Lenz et al. proposed an off-axis holography configuration that used one SLM to correct instrumentation and sample-induced aberrations [see Fig.2(d)]. They achieved a lateral resolution of 120 nm and an axial resolution of 173 nm when imaging tubulin at depths of 8?10 μm. In 2016, Patton et al. proposed an implementation that incorporates two AO elements to enable aberration correction in all three beam paths [see Fig.2(e)]. They used modal sensing with the Fourier ring correlation (FRC) metric and resolved glutamatergic vesicles in neural boutons in intact brains of Drosophila melanogaster at a depth of 10 μm. In 2019, Zdankowski et al. proposed an automated AO solution to correct instrumentation and sample-induced aberrations. They used modal sensing with the brightness metric and achieved super-resolution imaging of a 15 μm mitotic spindle with a resolution of 50 nm×50 nm×100 nm. On this basis, in 2020, Zdankowski et al. combined AO with image denoising algorithm by block-matching and collaborative three-dimensional (3D) filtering (BM3D) to enhance the image quality and super-resolved 3D imaging of axons in differentiated induced pluripotent stem cells growing under an 80 μm thick layer of tissue with lateral and axial resolutions of 204 and 310 nm, respectively [see Fig.3(a)]. In 2020, Antonello et al. proposed using wavelet analysis to quantify resolution loss and established a multivalued image quality metric. They achieved super-resolution imaging of CA1 pyramidal neurons in an organotypic hippocampal slice at a depth of 14 μm. In 2021, Hao et al. combined AO with 4Pi-STED. They used two deformable mirrors (DMs) in the two paths and analyzed the aberrations with modal sensing [see Fig.4]. They achieved sub 50 nm isotropic resolution of structures, such as neuronal synapses and ring canals previously inaccessible in tissues. Other indirect or direct wavefront detection techniques have been also used to measure aberrations in STED microscopy.In SIM [see Fig.6(a)], aberrations in excitation and emission paths decrease the imaging quality and should be corrected with AO. In 2008, Débarre et al. implemented sensorless AO in SIM. They investigated how the image formation process in this type of microscopy is affected by aberrations and performed aberration correction with modal sensing. In 2015, Thomas et al. combined sensorless AO with SIM and achieved super-resolution imaging of 100 nm fluorescence beads fixed beneath a C. elegans sample with a 140 nm resolution. In 2021, Zheng et al. proposed an AO correction method based on deep learning and utilized the method to correct aberrations with SLM, realizing super-resolution imaging of phalloidin-labeled actin in cultured BHK cells. Recently, direct wavefront sensing has also been used in SIM. In 2019, Turcotte et al. applied AO in SIM in vivo by generating the guide star with two-photon excitation as the input of the Shack-Hartmann wavefront sensor and performing aberration correction with a DM. They imaged the brains of live zebrafish larvae [see Fig.5(a)] and mice and observed the dynamics of dendrites and dendritic spines at nanoscale resolution. Similarly, in 2020, Li et al. used AO in optical-sectioning SIM [see Fig.6(b)] and achieved fast, high-resolution in-vivo imaging of mouse cortical neurons at depths of 21?29 μm [see Fig.5(b)] and zebrafish larval motor neurons at depths of 10?110 μm. In 2021, Lin et al. used direct wavefront sensing in SIM with a configuration that can be switched among wide-field imaging, structured illumination, and confocal illumination [see Fig.6(c)]. They used modal sensing to correct the aberrations of fluorescence beads and then recorded the image arrays in Shack-Hartmann wavefront sensor as a reference. Subsequently, they used confocal illumination to generate the guide star, input it into the Shack-Hartmann wavefront sensor, and reconstructed the wavefront. They decreased the peak-valley values of the wavefront amplitude from 1.5 to 0.1 μm when imaging C.elegans.In SMLM [see Fig.7(a)], aberrations in the emission path result in distorted PSFs and decreased localization precision, which should be corrected with AO. In 2015, Burke et al. proposed a technique for correcting aberrations using modal sensing with the sharpness metric [see Fig.7(c)]. They achieved a resolution of 78 nm laterally and 136 nm axially for microtubules at a depth of 6 μm. Tehrani et al. optimized aberrations using genetic algorithm [see Fig.7(d)] with the intensity-independent Fourier metric and increased the localization precision by four times at a depth of 50 μm. In 2017, Tehrani et al. proposed a real-time wavefront aberration correction approach based on particle swarm optimization [see Fig.7(e)] and the intensity-independent Fourier metric. They achieved a resolution of 146 nm for the central nervous system of Drosophila melanogaster at a depth of 100 μm. In 2018, Mlodzianoski et al. developed adaptive astigmatism using Nelder-Mead simplex algorithm to correct wavefront distortions with the weighted sharpness metric. They achieved a resolution of 20 nm laterally and 50 nm axially for mitochondria at a depth of 95 μm. In 2021, Siemons et al. proposed robust and effective adaptive optics in localization microscopy (REALM) combined with modal sensing using the weighted sharpness metric [see Fig.7(f)]. They achieved an FRC resolution of 76 nm for microtubules [see Fig.8(a)] and cytoskeletal spectrin of the axon initial segment at a depth of 50 μm. In 2023, Zhang et al. proposed deep-learning-driven adaptive optics (DL-AO) that examined aberrations from detected PSFs using deep neuron network [see Fig.7(g)]. They achieved a resolution of 14?31 nm laterally and 41?81 nm axially for mitochondria [see Fig.8(b)] and dendrites at a depth of 133 μm. In 2023, Park et al. developed closed-loop accumulation of single-scattering (CLASS), which measured complex tissue aberrations from intrinsic reflectance and performed compensation [see Fig.7(h)], and resolved subdiffraction morphologies of cilia and oligodendrocytes in entire zebrafish at a depth of 102 μm, improving the localization precision from 67 nm to 34 nm.Conclusions and ProspectsThis review summarizes the application of adaptive optics in super-resolution microscopy, including indirect and direct wavefront detection. Indirect wavefront sensing requires no setup modifications, except for inserting the wavefront correction elements, which is economical and practical. However, the low response speed and narrow dynamic range limit its effectiveness for severe distortions. Direct wavefront sensing can provide increased response speed and dynamic range, despite its increasing complexity in instrumentation. The future prospects of AO methods in super-resolution microscopy include increasing the field-of-view, response speed, and imaging depth. We expect that the AO method will be a general option in future super-resolution fluorescence microscopy.
SignificanceCells, the basic structural and functional units, play an essential role in the development, aging, disease, and death of organisms. Since the first microscopic observation of cells by Robert Hooke in 1665, numerous advanced technical and theoretical methods have been developed over the past centuries to microscopically visualize cells, enabling a thorough analysis of life activities from the cellular to the molecular levels. Cells are composed of numerous macromolecular complexes with different sizes and diverse compositions. For example, the eukaryotic 80S ribosome is composed of large and small subunits, and each subunit contains various ribosomal RNA (rRNA) and ribosomal proteins. These complexes are usually considered as core signaling hubs to precisely control cellular structures and functions during various biological activities. Accordingly, cells can properly generate immediate responses to frequent environmental changes and distinct cellular stresses. Therefore, a mechanistic investigation of the structural assembly of these key signaling hubs and their functional regulation is necessary to improve our understanding of life activities and to identify potential therapeutic targets for disease treatment.Currently, single-particle cryo-electron microscope (cryo-EM), which requires only a small number of samples for analysis and does not involve the use of crystals unlike traditional X-ray-based methods, is the most powerful tool in structural biology owing to its extremely high spatial resolution. However, precisely resolving a structure using cryo-EM involves purification or enrichment of the target biomolecules, which increases the risk of inconsistency between in vitro resolved structures and the native structures in cells. Notably, owing to the lack of molecular specificity, understanding the interactions among different molecules when resolving multi-component complexes is challenging. In addition, high-quality cryo-EM analyses depend on the computational averaging of thousands of images of identical particles with good homogeneity and are thus currently unsuitable for evaluating highly heterogeneous signaling hubs that determine cell fates.Excitingly, super-resolution microscope (SRM) has emerged as an effective solution to these above-mentioned challenges. Fluorescence imaging is an indispensable technical tool for modern biological research owing to its molecular specificity, in situ visualization feature, and multiplex analysis ability. Super-resolution imaging, which overcomes the optical diffraction limit, is an efficient method for visualizing the arrangement and functions of biological hundred-nanometer signalosomes at the subcellular scale or even with single-molecule precision.ProgressThis review first introduces the basic principles and technical development of several major types of SRM, including stimulated emission depletion microscope (STED), structured illumination microscope (SIM), photoactivated localization microscope (PALM), stochastic optical reconstruction microscope (STORM), point accumulation for imaging in nanoscale topography (PAINT), DNA-PAINT, and minimal photon fluxes (MINFLUX). These tools have facilitated precise visualization of various biological activities and targets at remarkably high temporal and/or spatial resolutions, even reaching the molecular or angstrom scale in some extreme cases (Figs.1-2). More importantly, to date, several representative SRM-based applications in life science research have been demonstrated. Through rationale optimization of the key steps in STORM (including structure preservation, fluorescence labeling, signal acquisition, and image analysis), Xin Chen’s group from Xiamen University first visualized the ordered organization of necrosomes at the nanoscale and revealed their underlying mechanism to effectively initiate MLKL (a mixed lineage kinase domain-like protein)-dependent necroptosis and to precisely control the transition between apoptosis and necroptosis in cells stimulated by tumor necrosis factor (TNF). Maria Pia Cosma’s group from the Barcelona Institute of Science and Technology employed single-molecule localization microscope (SMLM) to investigate genome organization, especially the formation of chromatin ring structures. They proposed that the transcription-dependent negative superhelix primarily drives the master molecule cohesin to generate ring structures in vivo. The team led by Ana J. Garcia-Saez from the University of Tübingend quantitatively imaged Bax- and Bak-mediated pores in the mitochondrial outer membrane during intrinsic cell apoptosis; they observed the interplay of apoptosis and inflammation by controlling the dynamics of the mitochondrial content release. Ardem Patapoutian’s group from the Scripps Research utilized the iPALM and MINFLUX technologies to directly visualize the conformational stages of the mechanosensitive channel PIEZO1 in complex cellular environments (Fig.3). Finally, based on the practical experience gained from our group’s efforts, we summarized some important strategies, such as methods to minimize reconstruction artifacts, improve labeling efficiency, and strengthen quantitative analysis of super-resolved images, to obtain high-quality super-resolution images using SMLM (Fig.4).Conclusions and ProspectsWith the groundbreaking innovation of SMLM in the past two decades, our understanding of structural organization and functional regulation in multiple types of cells undergoing various biological activities has improved. Although the classic genetic and biochemical experiments revealed the fundamental cellular mechanisms, cell imaging provides more precise and intuitive information on molecular interactions in situ. Therefore, the spatial resolution of SMLM can be further improved up to the molecular level to precisely depict an informative signaling network for a variety of critical biological processes. Thus, considering the continuous development of SMLM and other SRM technologies, we believe that in situ nanoscale functional organization of key signaling hubs will become one of the most promising research areas in cell biology in the near future. In addition, SMLM is expected to revolutionize research in science and technology and lead to outstanding discoveries in the next 5-10 years.
ObjectiveX-ray luminescence computed tomography (XLCT) technology uses X-ray excitation to stimulate specific luminescent materials at the nanoscale, termed phosphor nanoparticles (PNPs), to produce near-infrared light. Photodetectors then capture the emitted near-infrared light signals from these excited PNPs. Through suitable algorithms, the distribution of PNPs within biological tissues can be visualized. This method allows for structural and functional insights into biological tissues, showing great potential for advancement. There are two main types of XLCT systems: narrow-beam and cone-beam. The narrow-beam XLCT system exhibits higher spatial resolution, albeit at the cost of lower X-ray utilization efficiency. This inefficiency results in extended imaging times, limiting its potential for clinical use. Conversely, the cone-beam XLCT system improves X-ray efficiency and shortens detection time. However, the quality of the reconstructed images tends to be lower due to detection angle limitations. To overcome these challenges, there is a need for an innovative XLCT system that realizes rapid and highly sensitive data collection while also maximizing the use of X-ray technology. By addressing these issues, the clinical limitations of XLCT can be reduced to pave the way for its further development, thereby unlocking a plethora of possibilities.MethodsThis study introduces a new cone-beam XLCT system based on photon-counting measurements, complemented by an associated reconstruction method. Through the synergistic collaboration between the field-programmable gate array (FPGA) based sub-sampling unit and upper-level control unit, the system realizes automated multi-channel measurements. This integration shortens data acquisition time, boosts experimental efficiency, and mitigates the risks associated with X-ray exposure. After the completion of system implementation, we conduct experimental validation of the system and methodology. Specifically, a fabricated phantom is subjected to multi-angle projection measurements using the established system, and image reconstruction and evaluation are performed using the Tikhonov reconstruction algorithm.Results and DiscussionsThe results of the dual target phantom experiment indicate that under the conditions of a cylindrical phantom radius of 40 mm, target radius of 6 mm, and distance of 14 mm from the dual target phantom (Fig.2), the similarity coefficient (DICE) of the reconstructed image of the dual target phantom exceeds 50% under six-angle cone-beam X-ray irradiation. Furthermore, the system fidelity (SF) exceeds 0.7 (Table 1). In the phantom experiment of dual targets with different concentrations, the system proposed in this study effectively distinguishes dual targets with a mass concentration difference of more than 3 mg/mL. The DICE of the reconstruction image maintains over 50%, SF remains over 0.7, and reconstruction concentration error (RCE) is also over 0.7 (Table 2). These phantom experiment results confirm the good fidelity and resolution capability of the proposed system. Nevertheless, numerous factors potentially degrade the experimental outcomes, such as the attenuation and scattering of X-ray beams in the XLCT system, the physical and chemical composition of the target body, or even uneven concentration distribution. Additionally, artifacts appear in the reconstructed images. In the future, our research will focus on optimizing algorithms and reducing noise to enhance the application of cone-beam XLCT for in vivo experiments.ConclusionsThis study comprehensively considers the advantages and disadvantages of two imaging methods in XLCT and proposes a photon-counting-based multi-channel cone-beam XLCT system. The system automation for multi-angle measurements is realized via FPGA and host computer interaction. Specifically, multi-angle cone-beam irradiation reduces data acquisition time, while photon-counting measurement enhances the system sensitivity. Furthermore, a phantom experiment is conducted to validate the effectiveness and practicality of the proposed system and algorithm. The results demonstrate a significant reduction in data acquisition time and an improvement in the utilization of X-rays.
SignificanceCancer remains a major life-threatening disease worldwide, as reported by the World Health Organization (WHO). Surgery is the primary therapy for most solid tumors, with the ideal outcome relying on a balance between complete tumor removal and maximal preservation of surrounding normal tissue. Current clinical imaging modalities such as magnetic resonance imaging (MRI), computed tomography (CT), and positron emission tomography (PET) lack the resolution to accurately delineate tumor boundaries. The gold standard in clinics for detecting tumor boundaries and infiltration is the histopathological analysis of surgical specimens via hematoxylin and eosin (H&E) staining. However, the H&E staining workflow requires time-consuming tissue processing, including formalin fixation, paraffin embedding, and manual staining, often taking more than a day before diagnostic results are available to surgeons. Consequently, there is an urgent demand for new real-time microscopic imaging techniques that can be used intraoperatively to provide instant feedback during tumor surgery.Recent years have seen promising developments in label-free nonlinear imaging techniques for real-time tissue pathology in the operating room. These techniques include multiphoton fluorescence microscopy, optical coherence tomography (OCT), Raman microscopy, and harmonic microscopy, which can visualize tumor margins without exogenous labels. Among these, third harmonic generation (THG) combined with second harmonic generation (SHG) offers a unique, label-free subcellular-resolution assessment of fresh and unprocessed tissues. THG signals arise from nonlinear three-photon optical responses at cell-cell and cell-matrix interfaces (Fig. 1), effectively detecting proliferative cells and vasculatures, key hallmarks of tumor pathology. THG microscopy stands out by providing sub-cellular resolution, rich cellular and molecular information, and images of H&E quality. Additionally, using a single beam, complementary information from SHG, two-photon excited fluorescence (2PEF), and three-photon excited fluorescence (3PEF) can be simultaneously collected, visualizing extensive architectural and molecular details. These advantages position THG imaging as a highly promising technique for intraoperative determination of tumor margins.In this review, we explore the fundamental principles of the THG nonlinear process and discuss its latest applications in intraoperative tumor imaging. We highlight recent engineering innovations enabling miniaturized, portable THG imaging systems suitable for operating room deployment. We also review pioneering efforts in developing THG-capable endoscope probes using flexible fiber-optics, potentially integrating with standard surgical equipment. Embedding THG microscopy seamlessly into clinical workflows can provide surgeons with real-time, in-situ histopathology, enhancing surgical outcomes without disrupting the surgical rhythm. This review aims to accelerate the translation and adoption of label-free nonlinear optical imaging, particularly THG microscopy, as a valuable intraoperative guidance tool.ProgressRecent studies have demonstrated the potential of integrated THG, SHG, and multiphoton fluorescence microscopy for ex-vivo characterization of freshly resected human brain tumors (Fig. 2), ovarian tumors, breast cancer specimens, lung tumors (Fig. 3), and other tumor types. These studies reveal pathological hallmarks such as increased cellularity, nuclear pleomorphism, and vascular proliferation. The in-situ extraction of tumor pathological features underscores THG imaging's potential to improve surgical outcomes. Efforts are underway to transition THG microscopy from benchtop to clinically viable tools. Most THG microscopes are currently confined to research labs, with large volumes, complex opto-mechanical components, and limited consideration for patient safety or imaging stability. To facilitate widespread intraoperative use, miniaturized and portable THG imaging platforms are necessary. Researchers in the Netherlands and the USA have independently developed compact, multimodal THG microscopes, and these devices have been tested in clinical settings, such as operation rooms or pathological laboratories, for pilot clinical validation (Fig. 4). These devices enable on-site assessment of surgical specimens and provide rapid diagnostic feedback for tumor classification and margin determination, assisting surgeons in decision-making. However, existing miniaturized THG microscopes are limited to ex-vivo imaging. To enable real-time, in-situ guidance without tissue removal, endoscopic techniques are essential for THG imaging. The nonlinear imaging field is witnessing increasing efforts to design THG-capable endoscopes, drawing from innovations in 2PEF/3PEF, SHG, OCT, and Raman microscopy (Fig. 5). THG endoscopy is still in its early stages, presenting numerous opportunities for scientific research, technology translation, and clinical studies.Conclusions and ProspectsTHG imaging shows promise for real-time intraoperative assessment of various cancer types. Significant progress has been made in developing compact, portable THG imaging systems for intraoperative use. Currently, only two groups have begun clinical testing with their portable THG microscopes. More systematic clinical testing is needed to further mature this technology for routine operation room use. Additionally, technical translations from other imaging modalities are required to advance THG endoscopy solutions. Despite the vast potential of THG microscopy for real-time, non-destructive assessment of fresh tissue, more efforts from both the scientific and industrial sectors are imperative to promote the translation of THG microscopes from laboratories to clinical settings.
ObjectiveBrain tumors pose a significant threat to human health, and fully automatic magnetic resonance imaging (MRI) segmentation of brain tumors and their subregions is fundamental to their computer-aided clinical diagnosis. During brain MRI segmentation using deep learning networks, tumors occupy a small volume of medical images, have blurred boundaries, and may appear in any shape and location in the brain, presenting significant challenges to brain tumor segmentation tasks. In this study, the morphological and anatomical characteristics of brain tumors are integrated, and a UNet with a multimodal recombination module and scale cross attention (MR-SC-UNet) is proposed. In the MR-SC-UNet, a multitask segmentation framework is employed, and a multimodal feature recombination module is designed for segmenting different subregions, such as the whole tumor (WT), tumor core (TC), and enhancing tumor (ET). In addition, the learned weights are used to effectively integrate information from different modalities, thereby obtaining more targeted lesion features. This approach aligns with the idea that different MRI modalities highlight different subregions of brain tumor lesions.MethodsTo address the feature differences required for segmenting the different subregions of brain tumors, a segmentation framework was proposed in this study, which takes the segmentation task of three lesion regions as independent sub-tasks. In this framework, complementary and shared information among various modalities is fully considered, and a multimodal feature recombination module was designed to automatically learn the attention weights of each modality. The recombined features derived by integrating these learned attention weights with the traditionally extracted features are then input into the segmentation network. In the segmentation network, the module automatically learns the attention weights of each modality and recombines these weights with traditionally extracted features. By treating the segmentation tasks of the three lesion regions as independent subtasks, accurate segmentation of the gliomas is achieved, thereby addressing the problem of differing multimodal information requirements for different regions. To address the inability of a 3DUNet to fully extract global features and fuse multiscale information, a U-shaped network based on scale cross attention (SC-U-Net) was proposed. Specifically, a scale cross attention (SC) module was designed and incorporated into the deep skip connections of a 3DUNet. By leveraging the global modeling capability of the transformer model, SC extracts the global features of the image and fully integrates multiscale information.ResultsFigure 7 shows the results of the ablation experiments with different configurations of the SC module. When the SC module is added to the 3rd to 5th skip connections, the network achieves the best integration of deep multiscale features, thereby enhancing the feature extraction capability of the model. The average Dice coefficient of the three regions reaches 87.98%, and the mean 95% Hausdorff distance is 5.82 mm, thereby achieving optimal performance. Table 1 lists the ablation experimental results. The best results are obtained when the proposed MR and SC modules are used together, with the Dice coefficients for the three subregions increased by 1.34, 2.33, and 7.08 percentage points. Table 2 presents the comparison results of the six state-of-the-art methods, indicating superior performance in most metrics. Figures 8 and 9 show the segmentation visualization results, revealing that the improved model can more accurately identify the tumor tissue, resulting in smoother segmentation boundaries. Additionally, by integrating multiscale features, the model gains a larger receptive field, reducing the unreasonable segmentation caused by a single-scale and limited receptive field. Therefore, the segmentation results are closer to the annotated images with minimal false-positive regions.ConclusionIn this study, a deep learning network framework, MR-SC-UNet, is proposed and applied to glioma segmentation tasks. The test results on the BraTS2019 dataset show that the proposed method achieves average Dice scores of 91.13%, 87.46%, and 87.98% for the WT, TC, and ET regions, respectively, demonstrating its feasibility and effectiveness. In clinical applications, accurate tumor segmentation can significantly improve the capabilities of radiologists and neurosurgeons for disease assessment and provide a scientific basis for precise treatment planning and risk assessment of patients.
ObjectiveMedical image registration is a spatial transformation process that aligns and matches the specific spatial structures contained in two medical images. It has been applied in disease detection, surgical diagnosis and treatment, and other fields. Traditional medical image registration methods are slow and computationally expensive. In recent years, researchers have made significant breakthroughs in medical image registration research using deep learning methods. Deep learning methods have increased the registration speed by hundreds of times, with a registration accuracy comparable to those of traditional methods. However, most patients have complex pathological conditions and lesions grow quickly, resulting in significant differences in the images collected at different stages. Existing deep learning-based registration methods have low registration accuracy and poor generalization performance when used for medical images of large deformations. Therefore, a multi-scale constraint network (MC-Net) for large-deformation 3D medical image registration based on multi-scale constraints is proposed.MethodsWe propose a multi-scale constraint network (MC-Net) for large-deformation 3D medical image registration based on multi-scale constraints. Three multi-kernel (MK) modules are designed as parallel multi-channel and multi-convolution kernels for the encoder to accelerate the training speed. A convolutional block attention module (CBAM) is added to skip connections and enhance the ability to extract complex semantic information and fine-grained feature information from large-deformation images. In order to improve the registration accuracy, MC-Net combines multi-scale constrained loss functions to implement a layer-by-layer optimization strategy from low resolution to high resolution.Results and DiscussionsIn an experiment, three publicly available 3D datasets (OASIS, LPBA40, and Abdomen CT-CT, with two modalities) were used for registration research. The effectiveness of MC-Net was demonstrated through original experiments, traditional comparison methods, deep learning comparison methods, ablation experiments, and multi-core fusion experiments. Based on the registration results shown in Figs. 5 and 6, MC-Net performed well in the registration of the OASIS and LPBA40 brain datasets, as well as for the Abdomen CT-CT abdominal dataset. In the brain image comparison experiment, the LPBA40 brain dataset was compared with a traditional registration method (ANTs) and three deep learning registration methods (VoxelMorph, CycleMorph, and TransMorph) in the same experimental environment. It was found that MC-Net outperformed the other methods in terms of detail registration in brain regions and overall brain contour deformation. The abdominal image comparison experiment compared two traditional methods (ANTs and Elastix) and two deep learning methods (VoxelMorph and TransMorph). It was found that MC-Net had some shortcomings in organ generation and contour deformation, but had better registration performance than the other methods in terms of blank area size and individual organ deformation. The ablation experiment was conducted using the LPBA40 dataset. It demonstrated the different roles of the MK and CBAM modules in processing medical images in MC-Net, which helped to improve the registration accuracy. In addition, this article also discusses the computational complexity of MC-Net. For large target images such as medical images, this article discusses how a multi-kernel (MK) fusion module can be designed to effectively reduce the computational complexity.ConclusionsIn response to the low accuracy and poor generalization performance of current large-deformation image registration methods, this paper proposes a medical image registration network (MC-Net) based on multi-scale constraints, with LPBA40, OASIS, and Abdomen CT-CT medical image datasets used as research objects. Information loss can be avoided by designing CBAM modules in skip connections to enhance the ability to extract differential information from large-deformation images. In addition, considering the slow registration speed caused by the large number of parameters when processing large-deformation images, the MK module was designed with a parallel path large kernel convolution structure to improve the registration speed without affecting registration accuracy. When combined with the multi-scale constraint loss function proposed in this article, it iteratively optimizes the deformation fields at three scales from low resolution to high resolution to improve the registration accuracy. The experimental results show that compared with other methods, this method has improved registration accuracy, speed, and computational complexity. The good registration performances in three datasets with MRI and CT modalities demonstrate the generalization ability of our method. Subsequent research will focus on designing an adaptive adjustment module for multi-scale constrained loss function hyperparameters, in order to solve the problem of the time-consuming hyperparameter tuning needed for loss functions in experiments and improve the experimental efficiency. In summary, MC-Net has practical value in the registration of large-deformation images.
ObjectiveThe fundus is the only part of the human body where arteries, veins, and capillaries can be directly observed. Information on the vascular structure of the retina plays an important role in the diagnosis of fundus diseases and exhibits a close relationship with systemic vascular diseases such as diabetes, hypertension, and cardiovascular and cerebrovascular diseases. The accurate segmentation of blood vessels in retinal images can aid in analyzing the geometric parameters of retinal blood vessels and consequently evaluating systemic diseases. Deep learning algorithms have strong adaptability and generalization and have been widely used in fundus retinal blood vessel segmentation in recent years. Digital image processing technology based on deep learning can extract blood vessels from fundus images more quickly; however, the contrast of fundus images is mostly low at the boundary of blood vessels and microvasculature, and the extraction error of blood vessels is large. In particular, the microvasculature, which is similar in color to the background and has a smaller diameter, renders it more difficult to extract less vascular areas from the background. To solve this problem, this study improves the classical medical-image semantic segmentation U-Net. To effectively extract the spatial context information of color fundus images, a multiscale feature mixing and fusion module is designed to alleviate the limitations of local feature extraction by the convolution kernel. Moreover, to solve the problem of low contrast of the microvessels in color fundus images, a microvessel feature extraction auxiliary network is designed to facilitate the network in learning more detailed microvessel information and improve the performance of the network's blood vessel segmentation.MethodsA microvascular segmentation model of a parallel network based on U-Net (MPU-Net) was designed based on microvascular detail information loss and limitations of the convolution kernels. The U-Net network model was improved. First, the U-Net network was paralleled with an auxiliary network for microvascular feature extraction (Mic-Net). Microvascular labels on the gold standard images of fundus blood vessels were obtained via morphological processing, and they were used in the auxiliary network of microvascular feature extraction to learn microvascular feature information. Second, the main network was introduced in a multiscale feature shuffling and fusion module (MSF). Through learning, more receptive field characteristic information can be used to relieve the convolution kernels under space limitations. In contrast, a channel-mixing mechanism was used to increase the interaction between channels to better integrate the characteristics of different receptive field sizes and microvascular characteristics. MPU-Net comprised two parallel U-Net branches: the main and microvascular feature extraction auxiliary networks. The network that used the whole blood vessel label to calculate the loss function is the main network, whereas the Mic-Net used the microvessel label to calculate the loss function. Each network branch had one lesser layer of upper sampling on the U-Net architecture to reduce the loss of detail. A multiscale feature shuffle fusion module was introduced into the main network to alleviate the limitation of obtaining local information by convolution and to fuse microvessel feature information into the main network more effectively. In this study, a multi-scale feature-mixing fusion module MSF was designed. The module had two input features. The first was the encoder output feature, which contained more spatial details and exhibited a better expression ability on thick blood vessels. The other was the decoder feature or the microvascular feature output by the decoder in Mic-Net, which contained more high-level semantic information.Results and DiscussionsWe use three publicly available datasets—DRIVE, CHASE_DB1, and STARE—to validate the proposed MPU-Net. The comparison results (see Table 1, Table 2 and Table 3) show that the MPU-Net proposed in this study performs well in terms of accuracy. As presented in Table 1, for the DRIVE test set, the accuracy, sensitivity, specificity, and AUC of the proposed MPU-Net are 0.9710, 0.8243, 0.9853, and 0.9889, respectively. Compared with existing segmentation method (TDCAU-Net), MPU-Net obtains the highest accuracy, sensitivity, specificity, and AUC, which are improved by 0.0154, 0.0056, 0.0097, and 0.0094, respectively. Further, compared with DG-Net, which exhibits a better overall segmentation performance, MPU-Net increases the values by 0.0106, 0.0629, 0.0016, and 0.0043, respectively. These results indicate that the MPU-Net proposed in this study performs well on the DRIVE dataset and is beneficial for improving the vascular segmentation accuracy of the DRIVE dataset from the perspective of microvascular feature extraction and multi-scale feature wash-and-wash fusion. As presented in Table 2, for the CHASE_DB1 test set, the accuracy, sensitivity, specificity, and AUC of the proposed MPU-Net are 0.9764, 0.8593, 0.9844, and 0.9913, respectively. Compared with the existing segmentation method (TDCAU-Net), MPU-Net obtains the highest accuracy, sensitivity, and AUC, which are increased by 0.0026, 0.0350, 0.0035, respectively. Further, compared with ACCA-MLA-D-U-Net, which exhibits a better sensitivity performance, it increases the values by 0.0091, 0.0191, and 0.0039, respectively. These results show that MPU-Net has a better segmentation performance on the CHASE_DB1 datasets, although the performance of MPU-Net is slightly lower than that reported by Mao et al. on specificity, but 0.0352 and 0.0020 higher than that in sensitivity and AUC, respectively. As shown in Table 3, for the STARE test set, the proposed MPU-Net values are 0.9768, 0.7844, 0.9907, and 0.9905 for accuracy, sensitivity, specificity, and AUC, respectively. Compared with the existing segmentation method (LUVS-Net), MPU-Net obtains the highest accuracy, specificity, and AUC, which are increased by 0.0015, 0.0046, and 0.1718, respectively. Further, compared with CS2-Net, which had the best sensitivity performance, it increases the values by 0.0098, 0.0094, and 0.0030, respectively. These results show that the proposed MPU-Net is better than the existing mainstream methods in terms of accuracy, specificity, and AUC, but the performance in the sensitivity index is not sufficiently good. In addition, there is a certain gap compared with CS2-Net, but the other indicators are better than those of CS2-Net. This indicates that on the STARE dataset, the model algorithm is significantly affected by the imbalance of vascular pixels and background pixel samples, and will improve the specificity by sacrificing the sensitivity. However, from the perspective of the overall evaluation indices of the model, namely accuracy and AUC, the MPU-Net model exhibits better performance. Further, from the perspective of the overall segmentation performance, MPU-Net is superior to the existing mainstream methods on the STARE dataset. This proves that it is helpful for the overall segmentation performance on the STARE dataset from the perspective of microvascular feature extraction and multi-scale feature shuffling and fusion. From the analysis of the three datasets, MPU-Net is confirmed to be better than the existing mainstream methods in terms of the accuracy and AUC indicators, indicating that the proposed method is beneficial for improving the overall segmentation performance of the model and has a certain generalization ability. For both the DRIVE and CHASE_DB1 datasets, the sensitivity index is superior to existing mainstream methods, indicating that the MPU-Net model can further improve the segmentation sensitivity of blood vessels. Thus, this study effectively improves the vascular segmentation performance of color fundus images from the perspectives of microvascular feature extraction and multiscale feature mixing and fusion.ConclusionsIn this study, from the perspective of retinal vascular segmentation, microvascular lesions are found to have an important reference value for systemic vascular diseases diagnose. However, there are still certain difficulties in microvascular segmentation tasks. Therefore, in the vascular segmentation task, the shortcomings of deep convolutional neural network for microvascular segmentation are studied, and a parallel network microvascular segmentation model based on U-Net is proposed for vascular segmentation tasks. To alleviate the limitations of feature extraction of convolutional neural networks, a multiscale feature-shuffling fusion module is used to exploit the feature information extracted by the convolutional neural network, and the continuity of vascular segmentation is effectively improved by increasing the interaction between channels and combining spatial multiscale information. To alleviate the loss of detailed information during feature extraction caused by the pooling operation in the U-Net encoder, a microvascular feature extraction auxiliary network was proposed to further extract microvascular feature information. The test results for the DRIVE, CHASE_DB1, and STARE validation sets demonstrate that the proposed network can effectively improve the vascular segmentation performance compared with existing networks with better performance. In the future, further research should be conducted based on the auxiliary network of microvascular feature extraction to extract more refined and comprehensive microvascular features.
ObjectiveDiabetic retinopathy (DR) is one of the most common complications of diabetes and one of the main causes of irreversible vision impairment or permanent blindness among the working-age population. Early detection has been shown to slow the disease's progression and prevent vision loss. Fundus photography is a widely used modality for DR-related lesion identification and large-scale screening owing to its non-invasive and cost-effective characteristics. Ophthalmologists typically observe fundus lesions, including microaneurysms (MAs), hemorrhages (HEs), hard exudates (EXs), and soft exudates (SEs), in images to perform manual DR diagnosis and grading for all suspected patients. However, expert identification of these lesions is cumbersome, time consuming, and easily affected by individual expertise and clinical experience. With the increasing prevalence of DR, automated segmentation methods are urgently required to identify multiclass fundus lesions. Recently, deep-learning technology, which is represented by convolutional neural networks (CNNs) and Transformers, has progressed significantly in the domain of medical-image analysis and has become the mainstream technology for DR-related lesion segmentation. The most commonly used methods are semantic segmentation-oriented CNNs, Transformers, or their combinations. These deep-learning methods exhibit promising results in terms of both accuracy and efficiency. Nevertheless, CNN-based methods are inferior in terms of global-character contextual information owing to their intrinsically limited receptive field, whereas Transformer-based approaches exhibit low local inductive biases and subpar perception of multiscale feature dependencies. Whereas models combining CNNs with transformers exhibit clear advantages, they require the extraction of deep semantic characteristics and direct feature concatenation from the same feature level without fully considering the importance of concrete boundary information for small-lesion segmentation, thus resulting in inadequate feature interaction between adjacent layers and conflicts among different feature scales. Moreover, these methods only focus on a certain type of DR lesion and seldom delineate multitype lesions simultaneously, thereby hampering their practical clinical application.MethodsIn this study, we developed a novel progressive multifeature fusion network based on an encoder-decoder U-shaped structure, which we named PMFF-Net, to achieve accurate multiclass DR-related fundus lesion segmentation. The overall framework of the proposed PMFF-Net is shown in Fig. 1. It primarily comprises an encoder module embedding a hybrid Transformer (HT) module, a gradual characteristic fusion (GCF) module, a selective edge aggregation (SEA) module, a dynamic attention (DA) module, and a decoder module. For the encoder module, we sequentially cascaded four HT blocks to form four stages to excavate multiscale long-range features and local spatial information. For a fundus image I∈R(H×W×C) (with height H, weight W, and C channels) as the input, we first applied a convolutional stem with a convolutional layer and MaxPooling layer for patch partitioning, which resulted in N patches X with a convolutional layer. The resulting patches X were embedded into the image tokens E using a trainable linear projection, and we denoted the output of the convolutional stem as F0=E. Subsequently, the embedded tokens E were fed into the four encoder stages to generate hierarchical feature maps Fi∈RH2i+1×W2i+1×Ci,(i=1,2,3,4). The designed GCF module gradually aggregates the adjacent features of various scales under the guidance of high-level semantic cues to generate an enhanced feature representation FiGCF(i=2,3,4) in each layer, except for the first layer and the narrow semantic gaps between different levels of features. Subsequently, the presented DA module dynamically selects useful features and refine the merged characteristics to obtain consistent multiscale features Aii=2,3,4 using a dynamic learning algorithm. Meanwhile, the developed SEA module incorporates low-level boundary features F1 and high-level semantic feature information A3 and A4 to dynamically establish the association between lesion areas and edges, refine lesion boundary features, and recalibrate the lesion location. In the decoder module, we introduced a successive patch-expanding layer between adjacent resolution blocks to double the size of the feature map and halve the number of channels. Within each convolution block, a convolution layer was embedded to learn informative features. Finally, we applied a prediction head to obtain the lesion-segmentation probability map Y∈R(H×W×K), where K indicates the number of categories corresponding to the K-1 lesion map and background map.Results and DiscussionsWe used two publicly available DR datasets, i.e., IDRiD and DDR, to verify the proposed PMFF-Net. The comparison results (see Tables 1 and 2) show that our PMFF-Net performs better than the current state-of-the-art DR lesion-segmentation models on the two datasets, with mDice and mIoU values of approximately 45.11% and 33.39%, respectively, for predicting EX, HE, MA, and SE simultaneously on the IDRiD dataset; and mDice and mIoU values of 36.64% and 35.04%, respectively, on the DDR dataset. Specifically, compared with H2Former, our model achieves higher mDice and mIoU values by 3.94 percentage points and 3.28 percentage points, respectively, on the IDRiD dataset, and 4.55 percentage points and 4.69 percentage points higher values, respectively, compared with those of PMCNet. On the DDR dataset, our model achieves the best segmentation results, outperforming H2Former by 5.17 percentage points and 6.15 percentage points in terms of mDice and mIoU, respectively, and surpassing PMCNet by 6.36 percentage points and 7.43 percentage points, respectively. Meanwhile, our model can provide real-time DR-lesion analysis, with analysis times of approximately 34.74 and 38.48 ms per image on the IDRiD and DDR datasets, respectively. The visualized comparison results shown in Figs. 6 and 7 indicate that the results predicted by our model are more similar to the ground truth compared with those of other advanced methods. The cross-validation results across datasets presented in Tables 3 and 4 show that, compared with other advanced segmentation methods, our model offers better generalizability. The perfect segmentation performance of the developed PMFF-Net may be attributed to the ability of our HT module in capturing global context information and local spatial details, the GCF module gradually aggregating different levels of multiscale features through high-level semantic information guidance, the DA module eliminating irrelevant noise and enhancing DR-lesion discriminative feature identification, and the SEA block establishing a constraint between the DR-lesion region and boundary. Additionally, the effectiveness of the components of the proposed PMFF-Net was justified, including the HT, GCF, DA, and SEA modules, on the IDRiD dataset.ConclusionsIn this study, we developed a novel PMFF-Net for the simultaneous segmentation of four types of DR lesions in retinal fundus images. In the PMFF-Net, we constructed an HT module by elegantly integrating a CNN, multiscale channel attention, and Transformer to model the long-range global dependency of lesions and their local spatial features. The GCF module was designed to merge features from adjacent encoder layers progressively under the guidance of high-level semantic cues. We utilized a DA module to suppress irrelevant noisy interference and refine the fusion multiscale features from the GCF module dynamically. Furthermore, we incorporated an SEA module to emphasize lesion boundary contours and recalibrate lesion locations. Extensive experimental results on the IDRiD and DDR datasets show that our PMFF-Net perform better than other competitive segmentation methods. By performing cross-validation across datasets, the excellent generalizability of our model can be similarly demonstrated. Finally, we demonstrated the effectiveness and necessity of the proposed model via a comprehensive ablation analysis. The developed method can serve as a general segmentation framework and has been applied to segment other types of biomedical images.
ObjectiveFluorescence molecular tomography (FMT), which can observe the three-dimensional distribution of fluorescent probes in small animals via reconstruction algorithms, has become a promising imaging technology for preclinical studies. The strong scattering property of biological tissues and limited boundary measurements with noise have resulted in the FMT reconstruction problem being severely ill-posed. To solve the problem of FMT reconstruction, some studies have been conducted from different aspects, e.g., the improvement of forward modeling and many regularization-based algorithms. Owing to the ill-posed nature and sensitivity to noise of the inverse problem, it is a challenge to develop a robust algorithm that can accurately reconstruct the location and morphology of the fluorescence source. Traditional reconstruction algorithms use the l2 error norm, which amplifies the influence of noise and leads to poor reconstruction results.MethodsIn this study, we applied the Huber iterative hard threshold (HIHT) algorithm to fluorescence molecular tomography. The HIHT algorithm modifies the l2 norm cost function into a robust metric function, and the inverse problem is modeled as a constrained optimization problem that is combinatorial in nature. The robust metric function combines the l1 and l2 loss functions to vary the robustness and efficiency of the algorithm by setting a user-defined tuning constant. In the presence of noise, the HIHT algorithm can effectively reduce the influence of noise and enhance the robustness of the algorithm.Results and DiscussionsNumerous numerical simulations and in vivo mouse experiments are conducted to evaluate the performance of the HIHT algorithm. The reconstruction performance of the HIHT algorithm is illustrated by the contrast-to-noise ratio (CNR), Dice coefficient, location error (LE), normalized mean square error (NMSE), and time. Quantitative and qualitative analyses show that the HIHT algorithm achieves the best reconstruction results in terms of the localization accuracy, spatial resolution of the fluorescent source, and morphological recovery, compared with the FISTA, Homotopy, and IVTCG algorithms (Figs. 1, 4). To further verify the robustness of the HIHT algorithm, we perform four sets of experiments with different Poisson and Gaussian noise intensities (Fig. 2 and Fig. 3). As the noise intensity increases, the NMSE of the HIHT algorithm is always the smallest, indicating that it has the highest reconstruction accuracy. At the same noise intensity, the HIHT algorithm has the smallest LE, indicating that it reconstructs the target closest to the position of the real source. When the noise intensity increases, the Dice coefficient of the HIHT algorithm is higher than those of the other three algorithms, which indicates that the HIHT algorithm has a better morphological reconstruction ability. The CNR fluctuation of the HIHT algorithm is smaller than the CNR variations of the other three algorithms in the 10%?25% noise range. The results show that when the noise level is lower than 25%, the HIHT algorithm still obtains satisfactory reconstruction results, compared with those of the other three algorithms. To further evaluate the reconstruction performance of the HIHT algorithm in practical applications, we also perform in vivo mouse experiments. The experimental results show that the HIHT algorithm has the smallest position error as well as the highest Dice coefficient, and the fluorescent bead reconstructed by the HIHT algorithm is the closest to the real fluorescent bead in terms of morphology, which further demonstrates the feasibility and robustness of the HIHT algorithm (Fig. 5). The experimental results show that the HIHT algorithm not only achieves accurate fluorescence target reconstruction, but also improves the robustness to noise.ConclusionsThis study investigates the problem of insufficient algorithm robustness in FMT, and the HIHT algorithm reduces the impact of noise on the reconstruction performance by using the Huber loss function as the residual term. With the same noise intensity, compared with the other three algorithms, the HIHT algorithm obtains the smallest LE and NMSE values as well as the largest CNR and Dice coefficient values, indicating that the HIHT algorithm has the best reconstruction performance. As the noise intensity increases, the reconstruction performance of the HIHT algorithm outperforms the other three algorithms, and the performance is more superior in the Poisson noise test, which indicates that the HIHT algorithm has the best reconstruction accuracy and robustness. The experimental results are consistent with the theoretical description in Section 2. These results indicate that the HIHT algorithm is insensitive to noise and has good robustness. In summary, when the measurement data sets are disturbed by noise, unlike the algorithms based on the l2 norm residual term, the HIHT algorithm uses a robust loss function to reduce the influence of the noise. Therefore, the accuracy and robustness of the HIHT algorithm are significantly improved such that the position and shape of the fluorescence source can be reconstructed more accurately. Overall, the HIHT algorithm has the best robustness in the case of accurate reconstruction. Therefore, this study can promote the preclinical application of FMT.
ObjectiveDue to its economic advantages, convenience of use, and wide applicability, fluorescence fluctuation-based super-resolution microscopy has rapidly advanced in recent years and has garnered increased attention and application. Compared with other super-resolution imaging techniques, fluorescence fluctuation-based super-resolution microscopy offers lower system costs and is particularly suitable for imaging live cells, demonstrating exceptional performance in observing subcellular structures and monitoring dynamic processes. Specifically, variations in the fluorescence fluctuation characteristics significantly affect the quality of the super-resolution reconstructed images. Therefore, a systematic investigation of image quality under various fluorescence fluctuation conditions is crucial for identifying the most suitable super-resolution imaging approach. These fluorescence fluctuation conditions include parameters such as the number of image-acquisition frames, signal-to-noise ratio, bright-to-dark state probability, and bright-to-dark fluorescence intensity ratio, which directly affect image clarity, the signal-to-noise ratio, and accuracy. Thoroughly examining these conditions, we can effectively select and optimize the super-resolution imaging method that meet specific research requirements and experimental conditions.MethodsWe developed a fluorescence fluctuation-based super-resolution comprehensive imaging reconstruction platform using MATLAB. This platform integrates four super-resolution methods, namely, SOFI, MSSR, MUSICAL, and SPARCOM, and can simulate fluorescence fluctuation signals under different conditions while simultaneously applying multiple super-resolution methods to generate datasets. The platform also supports the import and reconstruction of experimental data and presents the reconstruction results clearly and intuitively on the platform interface, thus allowing users to conveniently compare the imaging results of different approaches. A comprehensive image-quality assessment is then conducted on these simulated datasets. This study used four sets of data under different fluorescence fluctuation conditions and quantitatively analyzed the quality of the reconstructed images generated by the four super-resolution algorithms using five evaluation parameters: the resolution-scaled Pearson coefficient (RSP), resolution-scaled error (RSE), relative error of strength (K), signal-to-noise ratio (SNR), and resolution (R). These five parameters were used to determine the image reconstruction consistency, reconstruction error, image reconstruction uniformity, SNR of the reconstructed images, and improvements in the reconstructed image resolution. In addition, to assess the quality of images reconstructed by the super-resolution algorithms more comprehensively and objectively, this study assigned specific weights to these five evaluation parameters and defined a comprehensive evaluation factor (CEF). The weights were determined based on the relative importance of each parameter in the super-resolution imaging technology to ensure the contribution of each parameter was accurately reflected. To facilitate a better comparison of the performances of the four super-resolution algorithms, this study integrated a multilayer perceptron model with a CEF and datasets generated under various fluorescence fluctuation conditions. The model can determine the super-resolution image reconstruction method that best performs under various fluorescence fluctuation conditions by learning and analyzing the performance of different algorithms and outputting an optimal algorithm selection. In short, this model considers different fluorescence fluctuation conditions as inputs and uses a comprehensive evaluation factor of the reconstructed results from various super-resolution algorithms as outputs.Results and DiscussionsUnder the fluorescence fluctuation super-resolution comprehensive imaging reconstruction platform, fluorescence signals under varying fluorescence fluctuation conditions were generated. Super-resolution algorithms were applied to reconstruct the datasets and calculate their CEF values; some simulation results are presented in Table 1. The SPARCOM method demonstrates the best performance in terms of resolution and denoising capability, achieving a spatial resolution of up to 44 nm. However, this method relies heavily on the sparsity of image sequences for super-resolution reconstruction and struggles to reconstruct images accurately when the bright-state probability of the fluorescence fluctuation signal is too high or the bright-dark ratio is too low. The MUSICAL method, which has lower resolution capabilities, offers superior denoising performance but exhibits poor image reconstruction consistency, uniformity, and a longer reconstruction time. The MSSR method has moderate resolution capabilities but exhibits superior image reconstruction consistency and uniformity and can be combined with other super-resolution algorithms to obtain higher-quality super-resolution images. Although the SOFI method has lower resolution and denoising capabilities, it exhibits good image reconstruction consistency and uniformity and exhibits a higher image reconstruction rate. A multi-layer perceptron model was constructed with fluorescence fluctuation characteristics as inputs and the CEF values of different algorithms as outputs. An analysis of the generated and evaluated datasets showed that the constructed model achieves an accuracy of 92.3%, indicating reliable classification and recognition capabilities and enabling intelligent selection of the most suitable super-resolution image reconstruction method under varying fluorescence fluctuation signal conditions.ConclusionsWe developed a comprehensive super-resolution image reconstruction platform using MATLAB, which implements signal generation and super-resolution image reconstruction functions under various fluorescence fluctuation conditions. The performances of multiple super-resolution algorithms across different fluorescence fluctuation scenarios were systematically evaluated. Leveraging of the dataset generated by the software platform enabled us to introduce a multi-layer perceptron model for intelligent algorithm selection. This in turn allowed for accurate classification and identification of the optimal super-resolution technique. This approach enhances research efficiency and assists researchers in selecting the most suitable fluorescence fluctuation method for various subcellular super-resolution imaging studies. The approach can further advance the application of fluorescence fluctuation-based super-resolution imaging techniques for efficient investigation of the ultrafine structures of various biological subcellular organelles.
SignificanceIn the dynamic field of biomedical photonics, simulating light transport in biological tissues has become a cornerstone for advancing medical diagnostics, therapeutic interventions, and understanding photobiological processes. This research area is crucial due to its potential to transform a wide range of biomedical applications. These include high-resolution medical imaging technologies, such as optical coherence tomography and fluorescence imaging, and innovative therapeutic approaches such as photodynamic therapy. These simulations provide detailed insights into the complex interactions between light and biological tissues, enhancing the precision of medical diagnostics, allowing for tailored light-based treatments for individual patients, and furthering our understanding of light-induced biological effects.Monte Carlo (MC) simulation methods are at the forefront of this field, noted for their unparalleled flexibility and accuracy in modeling the stochastic nature of photon transport through media with diverse optical properties. The MC approach excels at replicating the complex phenomena of absorption, scattering, reflection, and refraction that characterize light’s interaction with heterogeneous biological tissues. Its ability to theoretically achieve any desired level of precision establishes it as the gold standard for simulating complex tissue optics scenarios, providing a crucial benchmark for validating results from other modeling techniques.However, the practical use of MC simulations is significantly hindered by their high computational demands, which require extended periods to produce accurate results. This limitation not only affects the method’s efficiency but also presents a major barrier to its application in real-time or high-throughput settings. Consequently, there is a pressing need for innovative acceleration techniques that can reduce the computational load of MC simulations without sacrificing accuracy. Developing and implementing such strategies is essential to broaden the use and impact of photon transport simulations in biomedical research and clinical practice, facilitating quicker and more precise analyses that can advance medical science and improve patient care.ProgressIn recent years, the field of MC simulations for photon transport has witnessed significant advancements aimed at overcoming the computational intensity that characterizes traditional MC methods. These innovations have led to substantially faster simulations, enhancing the practical applicability of MC techniques in biomedical photonics. Advances in this area include algorithmic improvements, the adoption of parallel computing strategies, and the development of specialized hardware accelerators.Firstly, advancements in algorithms have led to the development of modified MC methods that maintain accuracy while significantly reducing computation times. Techniques, such as the baseline simulation method, adjust parameters, such as photon quantity and scattering characteristics, to accelerate the process. Perturbation MC methods introduce minor changes to existing simulations to evaluate the impact of alterations in optical properties without needing a complete re-simulation. Hybrid approaches merge traditional MC simulations with analytical calculations, such as the diffusion approximation, balancing speed with accuracy. Additionally, variance reduction techniques, such as importance sampling and path length trimming, have been crucial in minimizing statistical fluctuations, thereby enhancing the precision of the simulation outcomes.Secondly, the integration of parallel computing techniques represents a significant advancement. The use of multicore CPUs and GPUs for parallel processing has transformed the field, allowing multiple simulations to run simultaneously. This development has not only drastically reduced computation times but also alleviated constraints related to the complexity of tissue models. Since the introduction of GPU-accelerated MC simulations in 2009, there has been a noticeable increase in research activity in this domain, reflecting a growing preference for parallel computing among researchers. The scalability of these technologies enables MC simulations to be executed on computer clusters, providing vast potential for addressing large-scale and complex simulation tasks.Lastly, the design and implementation of specialized hardware for accelerating MC simulations have shown promising results, particularly in energy efficiency and performance within computation-constrained environments. Although the development pace of these dedicated hardware accelerators lags behind that of general-purpose processors, they represent a forward-thinking solution capable of supporting mobile monitoring and photonic control applications.These advancements in MC simulation techniques not only signify substantial progress in the field but also underscore the collaborative efforts of the global scientific community. Institutions in China, the United States, France, and Germany have made notable contributions. As these technologies continue to advance, they promise to further improve the accuracy, efficiency, and practical applicability of photon transport simulations in biomedical research and clinical settings.Conclusions and ProspectsThe advancements in acceleration techniques for MC simulations have effectively addressed the inherent limitations of classical MC methods, particularly their computational intensity, thus broadening their use in various areas of biomedical photonics. Accelerated algorithms, parallel computing strategies, and specialized hardware have each been crucial in improving the efficiency and feasibility of MC simulations for modeling light transport in biological tissues. These developments have not only enabled faster simulations but have also maintained, and in some instances improve, the accuracy and reliability of the results.Looking forward, the ongoing evolution of computing technologies and algorithms promises significant further advancements in MC simulation acceleration. The integration of artificial intelligence and machine learning could revolutionize, for example, could offer novel approaches to optimize simulation parameters and predict outcomes, reducing computational demands. Additionally, the growing availability of high-performance computing resources and cloud platforms is set to democratize advanced MC simulations, making them more accessible to researchers and clinicians globally. As the field advances, the key challenge will be balancing computational efficiency with accuracy to ensure that accelerated MC simulations remain a robust tool for examining the intricate interactions between light and biological tissues. The future of MC simulation in biomedical photonics is promising, poised to substantially enhance medical diagnostics, therapy planning, and our understanding of photobiological processes.
SignificanceStructured illumination microscopy (SIM) is a pivotal technique in super-resolution microscopy as it offers an innovative approach to enhance the spatial resolution exceedingly beyond that achievable by conventional optical microscopes. SIM harnesses the principle of structured illumination, where finely patterned light interacts with the specimen, thereby generating moiré fringes containing high-frequency information that is otherwise unaccessible owing to the diffraction limit.Achieving genuine super-resolution via SIM is involves intricate steps, including capturing numerous low-resolution images under an array of varied illumination patterns. Each of these images encapsulates a unique set of moiré patterns, which serve as the foundation for the subsequent computational reconstruction of a high-resolution image. Although effective, this methodology presents some challenges. Biological samples, owing to their inherent irregularities and varying tissue thicknesses, can result in considerable variability in the quality and consistency of the captured moiré patterns. This variability hinders the accurate reconstruction of high-resolution images. Additionally, systematic errors can further complicate the process, thus potentially introducing artifacts or resulting in the loss of crucial details in the final image.Furthermore, sample damage due to prolonged light exposure must be considered when acquiring multiple images. Hence, the number of images required must be minimized without compromising the quality of the super-resolution reconstruction. Determining the optimal balance between the number of images and the quality of the final image is key in applying SIM to sensitive biological samples.Image-processing algorithms are widely employed to mitigate the effect of excessive image pairs on imaging results. In addition to the classical algorithms, recently developed deep-learning algorithms offer promising solutions. Deep-learning algorithms can extract meaningful information from limited data and efficiently reconstruct images using neural networks. This approach enables high-quality super-resolution images to be acquired faster without necessitating numerous input images. Consequently, in SIM image reconstruction, satisfactory results can be achieved using fewer input images. Furthermore, deep-learning algorithms can effectively manage irregularities and variations in samples. By learning the structure and features of samples, these algorithms can better adapt to different types of samples, thus improving the robustness and accuracy of image reconstruction. This is particularly important when managing complex biological samples, which typically exhibit diversity and variability. Therefore, analyzing and summarizing the applications and effectiveness of deep learning in SIM systems is crucial.ProgressIn deep learning, the widely recognized efficient neural network models include the convolutional neural network (CNN), U-Net, and generative adversarial network (GAN). The CNN, which is renowned for its capacity to automatically discern patterns and features within intricate datasets, is particularly suitable for the task mentioned above. By undergoing rigorous training on a substantial corpus of SIM images, the CNN learns to infer missing information that would otherwise require an array of supplementary images to capture. This predictive prowess enables the algorithm to amend the aberrations induced by SIM mode adjustments, thus significantly improving the quality of the reconstructed images. Because of the strategic deployment of skip connections within U-Net, which ingeniously amalgamates information from both the deeper and shallower layers, the network can effectively preserve abundant details and information throughout the upsampling phase. Furthermore, the integration of deconvolution processes not only amplifies the dimensions of the output image but is also pivotal in enhancing U-Net’s exceptional performance and widespread acceptance within the biomedical sector. In the context of SIM reconstruction, harnessing U-Net to extract supplementary insights from available images allows the algorithm to construct high-resolution images from a minimal subset of input images, thereby considerably diminishing the likelihood of specimen damage. By employing U-Net, one can reconstruct a super-resolved image similar to those afforded by classical algorithms using only three captured images. Furthermore, the implementation of GANs has significantly augmented the capabilities of deep-learning algorithms in SIM image processing. GANs comprise two dueling neural networks—a generator and a discriminator—that operate in tandem to fabricate highly realistic images. The generator synthesizes the images, whereas the discriminator assesses their veracity. Similar to U-Net, GANs can reconstruct super-resolved images from three original images. However, GANs can generate data through adversarial learning, and when coupled with other architectures, they can achieve even better results.In summary, to enhance performance and generate high-resolution images from a minimal number of original images, various neural network models are synergistically combined. Finally, the application of deep learning in nonstriped and non-super-resolution SIM yields encouraging results, thereby further expanding the possibility of its applicability.Conclusions and ProspectsThe integration of deep-learning algorithms into SIM image processing significantly advances the microscopy field. It not only addresses the technical challenges associated with achieving super-resolution but also provides new possibilities for investigating the nanoscale world with unprecedented clarity and detail. As deep-learning algorithms continue to advance, we expect more sophisticated algorithms to emerge and thus transcend the current boundaries of super-resolution microscopy.
SignificanceThe application of deep neural networks to image segmentation is one of the most prevalent topics in medical imaging. As an initial step in computer-aided detection processes, medical image segmentation aims to identify contours or regions of interest within images, thereby providing valuable assistance to clinicians in image interpretation, surgical planning, and clinical decision-making. Deep neural networks, which leverage their powerful ability to learn complex image features, have demonstrated outstanding performance in medical image segmentation. However, the use of deep neural networks for medical image segmentation has two significant limitations. First, different medical imaging modalities and specific segmentation tasks exhibit diverse image characteristics, leading to the low generalization capabilities of deep neural networks, which are often tailored to specific tasks. Second, increasingly complex network architectures with notable segmentation efficacy demand significant amounts of annotated image data, particularly those that require laborious manual annotation by medical experts.With the rapid advancement of large-scale pretrained foundation models (LPFMs) in the field of artificial intelligence, an increasing number of tasks have achieved superior results through the fine-tuning of LPFMs. LPFMs are generic models trained on massive amounts of data and acquire foundational and versatile representational capabilities that can be transferred across different domains. Consequently, various downstream tasks can be easily fine-tuned using universal models. Considering the challenges in medical image segmentation, including low model generalization and difficulty in dataset acquisition, universal LPFMs are urgently needed in the field of medical image segmentation to facilitate breakthroughs in artificial intelligence applied to medical imaging.Since its introduction as a foundational large model in the field of natural image segmentation, the segment anything model (SAM) has been applied across various domains with remarkable results. Although SAM has demonstrated powerful capabilities in natural image segmentation, its direct application to medical image segmentation tasks has yielded less-than-satisfactory outcomes. This can be attributed to two main factors. First, the training datasets contain shortcomings. SAM lacks sufficient representation of medical images in its training data, and medical images often exhibit blurry edges, which differ significantly from the clear edges present in natural images. Second, the characteristics of SAM prompts play a crucial role in segmentation performance. Only by judiciously selecting prompt strategies can the full potential of SAM be realized.For these two reasons, significant efforts have been directed toward fine-tuning SAM, adapting SAM to three-dimensional (3D) medical datasets, expanding SAM functionalities, and optimizing prompting strategies. Comprehensive review articles have summarized these endeavors, such as the study by Zhang et al., which extensively outlined advancements in fine-tuning SAM, expanding its functionalities, optimizing prompting strategies, and distilling the challenges faced by SAM in the field of medical image segmentation. However, a systematic summary of methods for applying SAM to 3D medical datasets is lacking. Zhang et al. primarily elaborated on the fine-tuning of SAM, its application to 3D medical datasets, and related automatic prompting strategies. Nevertheless, as research on SAM deepens and its performance across various datasets improves, efforts in fine-tuning SAM, adapting it to 3D datasets, and optimizing prompting strategies have become more sophisticated. In addition, SAM has been extended to integrate semi-supervised learning methods and has been applied to novel directions such as interactive clinical healthcare. To summarize comprehensively the progress of SAM adaptation to medical image segmentation as well as to address existing challenges and provide directions for further research, a review that specifically focuses on the application of SAM to medical image segmentation is essential.ProgressThis study extensively reviewed more than one hundred articles focusing on the utilization of SAM for medical image segmentation. Initially, this study furnished an exhaustive exposition of the SAM architecture and delineated its direct application to medical image datasets (Table 1). Then, an in-depth analysis of SAM's adaptation to medical image segmentation was conducted, emphasizing innovative refinements in fine-tuning techniques, SAM's integration into 3D medical datasets, and its amalgamation with semi-supervised learning methodologies (Fig. 3) alongside other emerging avenues. Experimental evaluations on two proprietary medical image datasets validated the enhanced generalization capabilities of the large models after extensive data fine-tuning (Table 2). In addition, the study confirmed the effectiveness of combining SAM with semi-supervised networks in generating high-quality pseudo-labels, thereby augmenting the segmentation performance (Table 3). Finally, the study delved into the current limitations, identified areas requiring improvement, elucidated the challenges encountered in SAM's adaptation to medical image segmentation, and proposed future directions, including the construction of large-scale datasets, enhancement of multi-modal and multi-scale information processing, integration of SAM with semi-supervised network structures, and expansion of SAM's application in clinical settings.Conclusions and ProspectsSAM is progressively being established as a potent asset in the field of medical image segmentation. In summary, although the integration of SAM into medical image segmentation holds great promise, it continues to face many challenges. Addressing these challenges requires a more comprehensive investigation and more refined approach, thus paving the way for effective implementation and further evolution of large-scale models in the domain of medical segmentation.
SignificanceKnown for its non-invasive and non-destructive nature, optical microscopy can provide structural and functional insights into biological specimens, thus driving progress in fields such as biology, medicine, and related disciplines. Over the past four centuries, optical microscopy has witnessed significant developments. These have been particularly accelerated in the last century by technological advancements in lasers and computational methods. These advancements have led to revolutionary changes, making optical microscopy an essential tool in critical sectors such as healthcare, education, and food safety. With the increasing exploration in cellular biology and biomedicine, a growing need has arisen for optical microscopes with molecular or nanoscale spatial resolution, as exemplified by super-resolution optical microscopy (SRM). Of the various SRM techniques, stimulated emission depletion (STED) microscopy stands out because it achieves resolution enhancement by modulating the depletion power relative to the redshift in the excitation wavelength in the imaging setup. However, excessive power depletion poses challenges, including photobleaching of fluorophores and phototoxicity to biological specimens, which constrain the utility of STED in live-cell imaging scenarios. In recent years, researchers worldwide have collaborated to advance the field of STED microscopy with a particular focus on developing strategies to reduce depletion power. This effectively decreases the amount of power required for imaging while maintaining resolution accuracy. These studies are crucial for understanding the intricate details and underlying mechanisms in living organisms.ProgressIn this review, we discuss the basic principles of STED microscopy and emphasize its crucial role in achieving super-resolution imaging of biological samples. Achieving super-resolution imaging using STED microscopy requires precise control over the spatial, temporal, and spectral aspects (Fig. 1). By applying the theoretical framework that governs the resolution calculations in STED microscopy, we outline methods for achieving low-power STED microscopy from four key perspectives: optimizing STED probes, using single-molecule localization techniques, employing advanced image processing methods, and utilizing time-resolved detection approaches.We then provide a brief summary of the current nanoprobes designed for low-power STED imaging that encompass organic molecule dyes and organic and inorganic nanomaterials. Based on a comparative analysis of their performance parameters and imaging outcomes, we highlight the essential criteria for nanoprobes suitable for STED imaging, with a focus on attributes such as photobleaching resistance, low saturation intensity, and favorable biocompatibility. We also summarize and compare the imaging capabilities of STED microscopy and its derivative technologies. Noteworthy examples include MINFLUX, LocSTED, and MINSTED, which synergistically combine the strengths of STED and single-molecule localization microscopy (SMLM) to achieve substantial enhancements in imaging resolution (Fig. 2). In terms of image processing, we expound on the principles of differential image processing, explaining its effectiveness in modulating fluorescence signals across the spatial, temporal, and frequency domains to facilitate low-power STED imaging (Fig. 3). Moreover, by leveraging the insights into the relationship between the stimulated emission effect and fluorescence lifetimes, we advocate for the adoption of time-resolved detection modules to discern fluorescence photons with long lifetimes. Through techniques such as time-gated detection, phasor plot analysis, and ratiometric photon reassignment, we demonstrate the potential for enhanced resolution by selectively isolating photons with prolonged lifetimes (Fig. 4). Finally, we evaluate the prevailing challenges impeding the widespread adoption of low-power STED microscopy, emphasizing the need for future research endeavors that optimize image quality and enhance both imaging depth and the intelligence and automation of imaging systems. Our primary objective is to advance the application of STED microscopy, particularly in demanding domains such as thick-tissue imaging and in vivo investigations.Conclusions and ProspectsIn the field of super-resolution imaging, STED microscopy is a pioneering far-field technique distinguished by its real-time capabilities, ultra-high resolution, and three-dimensional layer-slicing capabilities. These attributes make STED microscopy highly promising for bioimaging applications. To extend the utility of STED microscopy to in vivo imaging scenarios, a primary objective is to effectively reduce the depletion power, which is a major focus for future advancements in STED microscopy. With continuing advancements in scientific technology and the increasing demand for various applications, low-power STED microscopy enhancements are anticipated to progress further. For example, tailoring imaging parameters to diverse experimental conditions can be facilitated by integrating artificial intelligence and machine learning methodologies. This facilitates automatic parameter matching and the identification and tracking of target structures, thereby mitigating the complexity associated with experimental operations and enhancing both imaging efficiency and accuracy. In addition, the integration of STED microscopy with complementary advanced technologies holds promise for realizing expanded capabilities, including large-depth, multicolor, and three-dimensional imaging. These advancements are expected to provide researchers in the fields of biology and medicine with powerful tools for understanding complex biological processes.
ObjectiveThree-dimensional (3D) tumor organoids, serving as in vitro models that replicate the critical structural and functional features of organs and tumor tissues, have demonstrated their unique value in disease modeling, personalized medicine, and drug screening. Patient-derived organoids (PDOs) not only recapitulate the morphological characteristics and physiological functions of their original tissues but also maintain the genetic and heterogeneity of tumors, rendering them invaluable resources for cancer research and treatment. However, current methods for analyzing organoid growth and drug effects have limitations, particularly in the absence of 3D high-throughput and label-free monitoring tools, hampering the more effective assessment of organoid growth and drug actions. To address this challenge, this study is dedicated to developing a comprehensive evaluation method based on optical coherence tomography (OCT) and machine learning algorithms. The aim is to establish a novel, non-invasive, label-free tool for the morphological characterization of organoids, enabling longitudinal evaluation of their responses to drug treatments. This approach holds significant potential for the application of PDOs in personalized cancer therapy, particularly for intrahepatic cholangiocarcinoma (iCCA), for which treatment options are limited.MethodsIn this study, we propose a method that combines OCT imaging with machine learning to perform longitudinal, accurate, label-free, and parallel morphological characterization of a large number of individual organoids within organoid clusters. Through 3D OCT imaging and organoid segmentation technology, we achieved 3D imaging and morphological analysis of individual organoids, including parameters such as organoid volume, organoid surface area, and organoid cavity volume. Subsequently, based on undersampling, we conducted a cluster analysis on multiple organoids within the organoid clusters to obtain statistical information on multi-dimensional morphological parameters for different categories. Feature selection and principal component analysis (PCA) were then applied to construct a comprehensive evaluation scoring function that combines the factor scores of each principal component and weights according to their variance contribution rates. Furthermore, we characterized the relative growth value of organoid clusters by calculating the difference in the comprehensive evaluation scores of the growth levels between two time points. Alternatively, the growth rate of the organoid clusters was represented by the slope of linear fitting based on the comprehensive evaluation scores from multiple time points. Ultimately, we validated the effectiveness of the comprehensive evaluation model of the growth levels based on the organoid clusters and PCA using adenosine triphosphate (ATP) testing results.Results and DiscussionsOur study results highlight the significant advantages of OCT imaging and machine learning in characterizing organoid growth and drug responses. A notable correlation is observed between organoid morphological changes and drug treatments, such as the transition of cystic organoids to solid organoids under the influence of medication (Fig. 3). The comprehensive evaluation model that we constructed shows an 82.9% consistency with traditional ATP biochemistry testing, which is a widely recognized indicator of cellular activity and proliferation (Table 5). More importantly, the correlation between the relative growth values derived from our comprehensive evaluation model and ATP measurements reaches an impressive 90.4%. This high degree of consistency confirms that our model can serve as a reliable proxy for assessing organoid growth and drug sensitivity. Additionally, the study results underscore the potential of our method to reveal morphological changes in organoids, which may be significant indicators of drug response and may provide new insights into the complexity of tumor-drug interactions.ConclusionsThis study marks significant progress in the field of organoid research and its implications for cancer treatment. By integrating OCT with machine learning, we have developed a robust and comprehensive evaluation model that is capable of accurately assessing organoid growth levels and responses to drugs. This method stands poised to revolutionize traditional approaches to drug efficacy screening and sensitivity testing, particularly for PDOs. The high consistency observed between our evaluation model and traditional ATP testing underscores its potential as a reliable and non-invasive tool in cancer research. As we transition into the era of personalized medicine, the precise measurement and prediction of individual organoid drug responses are becoming increasingly crucial. The methodology outlined in this study not only reveals the morphological changes of organoids under the influence of drugs but also lays the groundwork for a new technological platform for cancer drug screening and clinical drug sensitivity testing based on PDOs. Its aim is twofold: to deepen our understanding of tumor biology and to advance the development of more precise and effective cancer treatment strategies.
ObjectiveNeuroblastoma (NB) is a type of peripheral neuroblastic tumor commonly found in children and characterized by obvious heterogeneity in biological behavior and rapid development. Determining the differentiation type is helpful in assessing the prognosis of neuroblastoma for making early judgments regarding postoperative treatment options. Whole-slide images (WSIs) of the NB have ultrahigh resolution and contain rich information, facilitating clinical interpretation. However, early diagnosis is time-consuming and poses significant challenges. Considering the complex cellular environment and heterogeneity of NB, this study proposes a novel network, CSA-U-Net, for cell segmentation and classification of NB WSI. Additionally, a cross-pseudo-supervised (CPS) approach, combining different proportions of labeled and unlabeled data, is used for training, which improves the robustness and generalization ability of the model, thereby assisting pathologists in clinical diagnosis, reducing their workload, and decreasing the misdiagnosis rate.MethodsTo address the cell-level data labeling problem, this study adopt a deep learning method based on CPS, fully utilizing the distributional characteristics of unlabeled data and combining a small amount of labeled data, to improve the model’s generalization ability by having the two branches supervise each other. To address the complex cellular environment and heterogeneity of NB, channel and spatial attention modules are added to the bottleneck layer of U-Net network. The proposed novel network, CSA-U-Net, is served as the base network for the CPS model, effectively improving model accuracy. Finally, the K-means algorithm is used to classify and count poorly differentiated and differentiated NB cells in the pathology slide images. The percentage of differentiated NB to the total number of tumor cells is calculated, to assist pathologists in determining histopathological typing.Results and DiscussionsThe CPS approach for NB WSI segmentation is shown in Fig. 1, with CSA-U-Net as the underlying network for the two branches (Fig. 5). The CSA-U-Net network was compared with U-Net, DeepLabv3+, PSPNet, HrNet, SA-U-Net, HoVer-Net, and MEDIAR. The results showed that the CSA-U-Net outperforms the other methods in all indicators. The F1 score was 79.05% in poorly differentiated cells and 62.21% in differentiated cells, and the accuracy was 96.78%, which is an improvement compared with that of the traditional U-Net (Table 1). In the prediction result graph, the prediction results of CSA-U-Net exhibit more accuracy, clearer boundaries, and less noise in the image, relative to other networks. A lower error rate is observed in the regions prone to erroneous segmentation (Fig. 8). Next, the performance difference of the CPS method with CSA-U-Net as the base network, was explored for labeled to unlabeled data ratios of 1∶1, 1∶2, 1∶3, and 1∶4. The results show that the segmentation accuracy of the model gradually increases with an increase in the amount of unlabeled data, and the F1 score of the model improves faster before the ratio of labeled to unlabeled data reaches 1∶3. After the ratio reaches 1∶4, the model enhancement is slower, and the speed of accuracy enhancement decreases significantly (Table 2). Subsequently, the CPS method was compared with other semi-supervised methods, at a 1∶3 ratio of labeled to unlabeled data. The CPS method showed the best detection performance, with F1 score of 80.99% in poorly differentiated cells, 65.40% in differentiated cells, and 97.99% accuracy (Table 3). Finally, the different types of cells in the prediction results were counted using the k-means method and compared with the gold standard of physicians (Fig. 9). The average accuracy of the counting results of poorly differentiated and differentiated NB cells was 94.00% and 89.89%, respectively (Table 4). This result indicates that the method in this study excels in the counting accuracy of poorly differentiated and differentiated cells and operates stably in images of any size, further validating the reliability of the method.ConclusionsTo address the problem of large amounts of cellular data and heavy labeling in NB images, this study adopted a CPS approach for model training. By introducing unlabeled data during training, the model can better capture the features of poorly differentiated and differentiated cells, thereby more accurately extracting and categorizing these cells from tissue backgrounds and displaying better adaptation to the variability and complexity of different samples. The CPS approach ensures the consistency of the two branches in terms of network structure while making them differ in parameter space through different initializations and independent training, which drives the model to learn a more robust and comprehensive feature representation. Meanwhile, for the features of NB pathology slide images, this study proposes a CSA-U-Net network model, incorporating an attention mechanism based on the original U-Net network, which further improves the accuracies of the segmentation and classification results. This study is based on the CSA-U-Net network and effectively integrates labeled and unlabeled data using a CPS semi-supervised model. The experimental results show that the CSA-U-Net network exhibits better performance on the NB dataset than existing control methods, with the segmentation accuracy of the model gradually improving as the amount of unlabeled data increases, which further validates the effectiveness of the CPS method. Finally, the K-means method was used to count the different types of cells in the model prediction results for pathological staging. The method proposed in this study effectively reduced the workload of pathologists, improved diagnostic efficiency, and is of great significance in determining the prognosis of NB.
ObjectiveOwing to the increasing prevalence of oral diseases, the societal demand for oral medical diagnosis has augmented steadily. This has increased the workload for oral health professionals, thus imposing higher requirements on their expertise and diagnostic efficiency. The interpretation of oral panoramic films is crucial in evaluating the oral health of patients. However, professional dentists are scarce in China, and a large number of film readings can take up too much of the doctor’s diagnostic time. The advent of artificial intelligence technology has expanded its application in the medical field, particularly in medical image analysis, and has yielded favorable results. Currently, most studies focus on individual tooth diseases. However, patients typically present multiple oral lesions simultaneously, including dental caries, apical periodontitis, furcation involvement, and impacted teeth. Owing to the complexity of these diseases, the existing technologies cannot satisfy actual clinical requirements. This study aims to leverage deep learning to recognize image features by employing a deep-learning network model to promptly and accurately identify diseased areas in oral panoramic films. The goal is to provide comprehensive results regarding conditions such as caries, periodontal disease, impacted teeth, and missing teeth. This approach aims to facilitate doctors in promptly and accurately diagnosing conditions, thereby alleviating diagnostic pressure stemming from inadequate medical resources.MethodsIn this study, we propose an efficient disease-recognition network named YOLO-Teeth (You only look once-teeth), which is based on YOLOv5s, to identify caries, impacted teeth, periapical periodontitis, and bifurcated root lesions. To enhance the feature-extraction capability of the backbone network, the Triplet attention mechanism is introduced such that the network recognizes the symptoms more accurately. A BiFPN module is used in the neck region to achieve a complete integration of deep and shallow features, thus ensuring that the network can process complex information in the panorama more effectively. The CIoU loss function is replaced by the MPDIoU loss function to improve the positioning accuracy of the network.Results and DiscussionsBased on the data presented in Table 1 and Fig. 6, the Triplet attentional-mechanism module outperforms the other 5 attentional mechanisms when the dimensionality reduction method is used in the oral disease recognition model. YOLOv5s, which employs the Triplet attention mechanism, demonstrates the most stable detection performance across various disease targets, with minimal fluctuations in the recognition performance of four diseases. Additionally, the accuracy rate (P), recall rate (R), and mean average precision (PmAP) of the model increase to 79.9%, 79.6%, and 85.9%, respectively, thus demonstrating the best comprehensive evaluation effect. Table 2 shows that, compared with the YOLOv5s network, YOLO-Teeth shows higher values in terms of the P, R, and PmAP by 5.0%, 3.2%, and 4.1%, respectively. Furthermore, YOLO-Teeth exhibits clear advantages over other mainstream detection networks, as shown in Table 3.ConclusionsThe YOLO-Teeth network proposed in this study is an efficient disease-recognition network based on YOLOv5s. Its feature-extraction capability is enhanced by introducing the Triplet attention module, whereas the integration of deep and shallow feature layers is improved using the BiFPN module. The CIoU loss function is replaced by the MPDIoU loss function, thereby enhancing the accuracy of disease-location identification. Ablation and comparison experiments are conducted using an oral panoramic-disease dataset. Experimental results show that compared with the YOLOv5s network, YOLO-Teeth shows higher values in terms of the P, R, and PmAP by 5.0%, 3.2%, and 4.1%, respectively. YOLO-Teeth is clearly advantageous compared with other mainstream detection networks. Therefore, YOLO-Teeth is suitable for disease recognition in oral panoramic films. The current research disparity in obtaining comprehensive disease-recognition results is addressed in this study. The findings obtained enable doctors to diagnose diseases promptly and accurately, thereby alleviating diagnostic pressure.
ObjectivePolarization-sensitive optical coherence tomography (PSOCT) can noninvasively obtain depth-resolved optical tomographic images of biological tissue samples. PSOCT is based on OCT with the additional function of detecting changes in the polarization states of light backscattered from different depths after polarized light is incident on the sample. It can provide polarization-related information about a sample, such as the Stokes parameter, Jones and Mueller matrix, phase delay, and depolarization, and it can distinguish structural specificities that OCT contrast cannot. PSOCT can be applied to detect changes in the function, structure, and activity of human tissues, which has major application prospects in medical diagnosis. In conventional PSOCT, the interference signals from the interferometer output are measured using horizontal and vertical polarization channels, thus requiring two separate spectrometer cameras. This increases the size of the system, adds additional costs, and requires strict triggering of hardware and software to avoid any time delay between the signal acquisition of the two cameras under the phase-based PSOCT algorithm. It also requires high response consistency of the charge-coupled device (CCD). Accordingly, PSOCT technology that utilizes a single spectrometer is currently under development. Some existing PSOCT systems based on a single camera can achieve only time-sharing or real-time detection. In this work, a PS and intensity dual-channel OCT measurement method is presented that can realize both time-sharing and real-time detection, thus providing a new method for laboratories in the analysis of the polarization information of samples.MethodsIn this study, a theoretical model of a PSOCT system with dual reference arms for a single camera is first established. Based on the traditional spectral-domain OCT (SDOCT) system, an additional reference arm is introduced, where the function of the two reference arms is to provide a pair of orthogonally polarized lights. A neutral filter is added to the other two reference arms to attenuate the light intensity and to ensure that the intensity of the light returned by the reference arm does not exceed the CCD acquisition threshold. A linear polarizer is added to the light source exit module, and four manual polarization controllers are added to the optical path. The system can realize the conversion between SDOCT and PSOCT imaging. A dual-reference arm detection system based on a single-mode fiber and polarization controller is then constructed. The quarter-wave plate in the existing SDOCT system is replaced with a single-mode fiber and polarization controller. The polarization module function is then expanded, and micron longitudinal high-resolution imaging performance is achieved using a broadband light source. The signals from the horizontal and vertical channels are collected using a single camera in real time. A Jones matrix model of the system is established, and the obtained tomography images are processed by combining the Stokes parameters. Relative reflectance, phase delay, and depolarization information are obtained to reconstruct the intensity, phase delay, and depolarization maps of the in vitro biological tissues, further verifying the feasibility of the proposed PS and intensity dual-channel OCT method as well as the imaging capabilities of the established system.Results and DiscussionsAn OCT system is established to realize the conversion of PSOCT and SDOCT as well as the functions of single-channel time-sharing detection and dual-channel real-time detection (Fig.1). A polarization characteristic model of the system is developed, and the calculation method for the sample polarization parameters is analyzed. Results show that a cross-sectional strength diagram of the sample tissue is obtained by processing the signals detected from the two orthogonal channels. The collected signal strength is then solved along the depth direction, and the defect polarization information is extracted using the Jones matrix and Stokes operation. The birefringence and depolarization parameters of the biological tissue as well as images of the polarization parameters of the biological tissue samples are obtained. The strength, phase delay, and depolarization of the sample beef (Fig.2) and chicken (Fig.4) tendons are reconstructed using time-sharing and real-time detection methods, demonstrating the feasibility of the system scheme and verifying its dual-channel detection capabilities.ConclusionsThe polarization characteristics of biological tissues can reveal the unique characteristics of biological tissues that cannot be described by isotropic properties, including tissue birefringence and depolarization. Depth-resolved isotropic intensity images and anisotropic polarization parameter images of biological tissue samples can be reconstructed noninvasively using a micron-resolution spectral domain PSOCT system. In this study, a dual-channel PSOCT system is proposed. The system is based on an optical fiber and single camera and has an imaging rate of 104 frame/s. An ultra-wideband light source, large numerical aperture objective lens, and common optical path structure are used to achieve horizontal and vertical resolutions at the micron level, ensuring the detection of tiny structures in tissue samples. The signals of the two channels can be collected at different moments such that horizontally and vertically polarized light can be incident on the same spectrometer at adjacent time to realize time-sharing detection. The interference signals of the two orthogonal channels can be collected simultaneously to achieve real-time detection. The images of the two channels are separated on both sides of the CCD, and the CCD can simultaneously contain the spectral interferograms formed by the horizontal and vertically polarized beam components. The proposed polarization imaging system provides a new method for the practical application of PSOCT and SDOCT in clinical diagnosis.
ObjectiveFull-field optical coherence tomography (FFOCT) is a contactless, high-resolution, real-time imaging method based on OCT technology. It utilizes the backscattering ability of the internal structure of the tissue to obtain a light signal with structural information about the tissue through the detector and then restores the real internal structure through a computer. With the introduction of the Linnik interferometer structure with two identical large numerical aperture microscope objectives into the FFOCT system, the imaging resolution of the system has been improved to the submicron level. Therefore, scholars have begun to conduct further research on aspects such as system effectiveness, stability, practicality, cost, and efficiency. Because the reference and sample arms use two identical microscope objectives, their alignment accuracy is at the micron level, which cannot meet the imaging conditions by the constraints of the mechanical structure. The presence of these two microscope objectives also increases the cost of the system. For different application scenarios, both objectives should be replaced and recalibrated simultaneously, which is a cumbersome process. In this study, we built an FFOCT system based on a Mirau interference structure, integrated the reference arm in the original structure into a self-made Mirau attachment, and assembled it between the objective lens of the sample arm and the sample. This makes the system more compact and stable and reduces costs. The microscope objective at different magnifications can be replaced according to requirements, and different media can be used for index matching.MethodsBased on the imaging principle of the FFOCT system, we first simulated the Mirau interference structure using the optical simulation software VirtualLab Fusion. In the preliminary verification stage, only the Mirau-attachment structure was designed; thus, the microscope objective was simplified to an ideal lens that satisfied the software parameters. The feasibility of the solution was verified through light-field tracing and imaging experiments on the test surfaces at different positions. Based on the parameters obtained from the simulation, we designed and processed the mechanical structure, which included two pieces of glass with reflective and beam-splitting functions as well as a structure for the adjustment and filling of the medium. The obtained structure was assembled into a system for imaging experiments, and a USAF standard-resolution target and plane mirror were imaged to measure the lateral and axial resolutions of the system, respectively. Using PCB, onions, and plant leaves as imaging objects, a four-step phase-shift method was employed as the solution method. The imaging results were used to verify the tomographic capabilities of the system.Results and DiscussionsThe feasibility of this principle is verified through simulations. The light-field tracing diagram conforms to the imaging principle, and the interference fringe pattern obtained by imaging the test surface at different positions is also in line with expectations (Fig. 2). To meet the actual conditions, we attached the reflector to a piece of glass with the same material and thickness as the beam splitter, adjusted the parameters, and performed simulations again. The results obtained still meet expectations (Fig. 3). A Mirau attachment was designed and fabricated (Fig. 4). Based on optical principles, the theoretical lateral and axial resolutions are calculated to be 1.73 and 7.56 μm, respectively. By imaging the USAF standard-resolution test target, the actual measured value of the lateral resolution is determined to be 2.19 μm (Fig. 5). An interference fringe intensity distribution diagram is obtained by imaging the plane mirror. Based on the resulting diagram, the actual measured value of the axial resolution is calculated to be 9.1 μm (Fig. 6). A certain position on the PCB contains two structures with different heights. The height difference is greater than the coherence length of the light source. When interference fringes appear on one side, no interference fringes appear on the other. Therefore, the calculated image contains only one side of the structural information, reflecting the tomographic capability of the system (Fig. 7). When imaging biological samples, their internal structures are restored. As the depth increases, the structural morphology changes and tomographic features are observed (Figs. 8?9).ConclusionsAn FFOCT system based on the Mirau interferometer is developed. This structure is more compact and stable than an FFOCT system based on a Linnik interferometer. The previous cross structure was changed to the current T-shaped structure, and the system complexity was reduced. The previously used FFOCT system uses two identical water-immersion objectives; however, this system requires only one microscope objective and does not require a water-immersion objective, which reduces system costs. Using a self-made Mirau interference objective structure, the parameters can be adjusted according to different requirements, and the microscope objective magnification and filling medium can be changed according to the imaging object. The tomographic capabilities of the system are verified by imaging abiotic and biological samples, and imaging with cell-level resolution is achieved. However, the resolution and imaging quality of the system are affected by the presence of environmental noise and noise generated by the operation of the system itself. For the four-step phase-shift method used in this system, mechanical vibrations cause a deviation between the actual phase shift and the set value, resulting in insufficient signal demodulation, thus retaining some interference fringes in the resulting image and affecting the imaging effect. Moreover, because the microscope objective is a commercial version and cannot be directly simplified into a lens, the self-made Mirau interference objective structure causes certain aberrations that affect the imaging quality and depth.
ObjectiveExisting deep learning-based methods for fluorescence image super-resolution can be broadly classified into two categories: those guided by peak signal-to-noise ratio (PSNR) and those guided by perceptual considerations. The former tends to produce excessively smoothed prediction results while the latter mitigates the over smoothing issue considerably; however, both categories overlook the ill-posed nature of the super-resolution task. This study proposes a fluorescence image super-resolution method based on flow models capable of reconstructing multiple realistic super-resolution images that align with the ill-posed nature of super-resolution tasks. Moreover, microscopy imaging is conducted in continuous time sequences naturally containing temporal information. However, current methods often focus solely on individual image frames for super-resolution reconstruction, completely disregarding the temporal information between adjacent frames. Additionally, structures in similar biological samples exhibit a certain degree of similarity, and fluorescence images collected possess internal self-similarity in the spatial domain. To fully leverage the temporal and spatial information present in fluorescence images, this study proposes a frequency- and spatial-domain joint attention module. This module aims to focus on features that significantly contribute to the prediction results, obtaining more accurate reconstruction outcomes.Similar to most supervised learning methods, our approach has a limitation in that it requires labeled paired image sets for training the network model. Generalization performance may significantly decline when applying the model to a test set with a distribution different from the training set. Acquiring labeled paired training data is not always feasible in practical applications. Therefore, future work may need to address the challenge of cross-dataset super-resolution reconstruction, considering optimization strategies and network improvements from a domain adaptation perspective.MethodsThis study introduces a flow-model-based multi-frame dual-domain attention flow network. Given a low-resolution image, the network learns the distribution of super-resolution images using flow models, enabling the reconstruction of multiple realistic super-resolution images to address the underdetermined nature of super-resolution tasks. Additionally, as the imaging process is typically continuous, the acquired raw data from a microscope has temporal relationships with adjacent frames. However, existing deep learning-based fluorescence image super-resolution methods often neglect the temporal priors present in multiple input frames over time. Moreover, biological sample structures exhibit internal self-similarity. Therefore, by constructing a frequency-domain and spatial-domain joint attention module, the network is guided to focus extensively on features that contribute significantly to the prediction results, further enhancing the network’s performance. The proposed flow-model-based multi-frame dual-domain attention flow network consists of a flow model and a frequency-domain and spatial-domain joint attention module. The flow model, composed of multiple reversible modules, facilitates a reversible mapping between the target and latent space distribution. The frequency-domain and spatial-domain joint attention module achieves conditional feature extraction and includes a set of frequency- and spatial-domain attention blocks. These blocks comprise Fourier channel attention blocks, spatial attention blocks, and convolutional layers, serving to extract temporal, spatial, and aggregated features from the fluorescence image, respectively. Furthermore, the study employs skip connections to enable feature reuse and prevent gradient vanishing.Results and DiscussionsThis study demonstrates the importance of temporal information by comparing the proposed method, a multi-frame dual-domain attention flow network (MDAFN), with a single-frame dual-domain attention flow network (SDAFN). Quantitative evaluation metrics include PSNR and learned perceptual image patch similarity (LPIPS). Experimental results indicate that the MDAFN outperforms the SDAFN, and the indexes of PSNR and LPIPS on the three data sets are shown in Table 1. Moreover, visually, the images reconstructed using the MDAFN exhibit improvement over those generated using the SDAFN (Figs. 7?9). Finally, a comparison between the proposed method and state-of-the-art super-resolution reconstruction methods is presented. The results indicate that when the standard deviation of hyperparameters is set to zero, the PSNR of the super-resolved images obtained using the proposed method is comparable or even superior to that obtained using other methods. For LPIPS, the proposed method outperforms other methods. When the standard deviation is greater than zero, the LPIPS obtained using the proposed method is further decreased across the three datasets (Table 2). The reconstructed results using the proposed method visually resemble the ground-truth images more closely. In contrast, other methods generate over-smoothed, signal-depleted, or artificially enhanced reconstructions with poorer subjective quality (Figs. 10?13).ConclusionsThis study proposes an MDAFN for high-quality super-resolution reconstruction of fluorescence images. Unlike conventional neural networks that directly learn deterministic mapping functions, our approach can predict various possible super-resolution images for a given low-resolution wide-field image, addressing the underdetermined nature of the one-to-many relationship in super-resolution tasks. Additionally, considering the high internal self-similarity of structures in similar live cell samples in both temporal and spatial dimensions of fluorescence images, we further introduce a frequency- and spatial-domain joint attention module based on multi-temporal input. This module aims to focus more on features contributing significantly to the prediction results, yielding more accurate predictions. Experimental results demonstrate that the proposed method outperforms other methods in terms of super-resolution image quality.
SignificanceAntibodies have potential for use in clinical diagnostics, therapeutics, and biomedical research. Since the approval of the first monoclonal antibody-associated drug by the United States Food and Drug Administration (FDA) in 1986, monoclonal antibodies have been considered the most promising targeted agents against diseases. Over the past four decades, an increasing number of monoclonal antibodies have been used in clinical settings and preclinical trials. Examples include daratumumab, which targets CD38 in multiple myeloma cells, and nivolumab, which reactivates T-cell recognition ability by inhibiting the PD-1/PD-L1 pathway and preventing cancer progression. However, large molecular monoclonal antibodies (molecular mass of ~150 kDa) have complex structures and physicochemical properties, which lead to in vivo limitations such as low tissue penetration, extended circulation, and slow clearance. In addition, high-price production and potential immunogenicity issues imped the development and application of monoclonal antibody-based imaging and therapeutic agents.A single domain antibody is a small antibody fragment first discovered in Camelidae, which can effectively recognize antigens of interest. Compared with conventional monoclonal antibodies, single domain antibodies exhibit superior properties, such as small size (~15 kDa), high affinity, high stability, low immunogenicity, and ease of tissue penetration, making them promising candidates for targeted imaging and drug delivery in research and clinical applications. This review systematically introduces progress in the research and application of single domain antibody-based probes in live-cell imaging, super-resolution imaging, in vivo fluorescence, and nuclear imaging. This review also discusses the future opportunities and challenges for single-domain antibodies in biomedicine.ProgressThe applications of single domain antibodies in cell imaging, including live-cell imaging and super-resolution imaging, were first introduced. Rothbauer et al. first investigated the construction of a chromobody in 2006 by fusing the gene sequences of a single domain antibody and a fluorescent protein, enabling its expression in live cells and binding to antigens of interest (Fig. 2). Subsequently, various chromobodies that target finer subcellular structures, such as organelle membrane actin, nuclear actin, and vimentin, have been developed. To improve the controllability and applicability of single domain antibodies in complex cellular environments, O'shea et al. developed a photocaged probe (Fig. 3). For super-resolution imaging, single-domain antibodies can be used to label subcellular structures with high densities and minimal linkage errors. The single domain antibody probes and labeling strategies for single molecule localization microscopy (SMLM), stochastic optical reconstruction microscopy (STORM), stimulated emission depletion microscopy (STED), and expansion microscopy (ExM) are summarized in the second section (Figs. 4 and 5). Jungmann et al. introduced a DNA barcoding method called resolution enhancement by sequential imaging (RESI), which improves the resolution of fluorescence microscopy to the Ångström scale using single domain antibodies (Fig. 6).In the third section, the bioimaging applications of single domain antibody probes are presented to demonstrate their advantages, including short circulation time, high affinity, deep tissue penetration, and rapid enrichment. Single domain antibodies conjugated with near-infrared fluorescent dyes have become effective for tumor imaging and image-guided surgery (Fig. 7). When combined with short half-life nuclides, such as 68Ga, 18F, and 64Cu, single domain antibody-based tracers demonstrate high specificity and low radiation risk in immunoPET and immunoSPECT imaging, which are better than the traditional 18F-FDG (Fig. 8).Conclusions and ProspectsThe advantages of single domain antibodies, including their small size, high stability, deep penetration, renal clearance, and low immunogenicity, are promising for biomedical applications. In cell imaging, single domain antibodies can be effectively modified using various tags, dyes, and adapters, making them versatile tools for live-cell and super-resolution imaging. At the organismal level, single domain antibodies exhibit a short circulation time, rapid clearance, and low risk of toxicity and immunogenicity, which facilitate real-time and highly specific in vivo imaging. Current preclinical data indicate that single domain antibody-associated probes have a tremendous translational potential. The first radiolabeled single domain antibody probe, 2Rs15d (HER2-targeting), labeled with either 68Ga or 131I, has entered clinical phase I trials.Numerous studies have been conducted on the design and screening of single domain antibodies that target various antigens. However, it is crucial to develop novel and effective biomarkers and screen for the corresponding high-specific single domain antibodies for the diagnosis and treatment of complex diseases, particularly heterogeneous tumors. The key to designing single domain antibody probes is exploring stable, universal, and controllable strategies for modification using dyes, chelators, and other labels. Because of the small size and limited active sites of single domain antibodies, the effects of conjugation sites, labeling methods, and properties of tags on single domain antibodies are significantly more pronounced than those on monoclonal antibodies. Although random labeling methods that rely on endogenous cysteine or lysine residues in proteins are simple and widely used, they may form heterogeneous products with variable functionalization ratios, and sometimes lead to a loss of targeting ability. Enzymes, including sortase-and microbial transglutaminase-mediated strategies and click chemistry, may be effective and promising approaches for constructing single domain antibody-based probes and their clinical translation. However, challenges remain regarding their application and improvement. In addition, advancements in chelators, isotopes, fluorescent dyes, imaging techniques, and other fields will continue to promote the development and translation of single domain antibody probes.
SignificanceOptical coherence tomography (OCT) technology was proposed in 1991. Based on the principle of low-coherence light interference, OCT features high resolution and a high signal-to-noise ratio; it is also non-invasive, non-destructive, and capable of three-dimensional imaging. It enables the imaging of microstructures within biological tissues, with significant application potential in fields such as biology, medicine, and materials. Consequently, it stands as one of the focal points in modern optical imaging research.The resolution plays a crucial role in the assessment of OCT because it directly impacts the image quality, thereby influencing the accuracy of disease diagnosis and condition assessment by medical professionals. Therefore, the pursuit of high resolution has been one of the primary directions in the development of OCT technology. Vis-OCT, which utilizes visible light with shorter wavelengths for imaging, offers higher lateral and axial resolution, enabling a finer depiction of microstructures within biological tissues.In comparison to NIR-OCT, which is commonly used in hospitals, Vis-OCT not only provides higher image resolution but also yields images with higher contrast, thereby revealing more information about biological tissues. The high resolution and contrast of Vis-OCT make it promising for widespread applications in fields such as ophthalmology and dermatology. By providing doctors with more detailed and precise image information, it facilitates more accurate observations and diagnoses of pathological tissues. Additionally, Vis-OCT can quantify sO2 through spectral analysis, providing a powerful tool for assessing retinal blood circulation and oxygen metabolism. Lastly, because the wavelength of visible light falls within the sensing spectrum of the retina, there may be a more direct relationship between the structure and function of the retina that can be observed by Vis-OCT, making these advantages worthy of further research.In practical applications, researchers have already conducted studies using Vis-OCT for various diseases. They have validated its ability to diagnose diseases such as glaucoma, macular lesions, and diabetic retinopathy and have identified some early biomarkers of diseases. This facilitates early intervention and treatment by medical professionals, thus providing better treatment outcomes and vision protection for patients.This paper provides an overview of Vis-OCT, introducing its technical characteristics and current applications and discussing future development trends. This will allow readers to better understand its advantages in medical imaging and recognize its value and significance in medical practice.ProgressThis paper reviews the progress of Vis-OCT research in the areas of system structure optimization, functional imaging research, animal imaging research, and clinical applications.In terms of system structure and functional imaging research, the article discusses the advantages and disadvantages of spatial coupling and couplers, along with related work. Additionally, in spectral analyzer optimization, the introduction of prisms achieves K-domain linearization, reducing errors in data processing, and the use of dual spectrometers improves the signal-to-noise ratio of the system. Furthermore, modulating the reference light path doubles the total imaging depth range and corrects the retinal curvature of wide-field imaging. This article also introduces the principles of DOCT, OCTA, and sO2 detection.Regarding mouse retinal imaging, this article describes the use of Vis-OCT for sO2 detection in mouse retinas and the quantification of the mouse retinal oxygen metabolism rate. Additionally, this paper shows how researchers use Vis-OCT to observe the effects of prematurity on mouse retinal lesions, dexamethasone-induced high intraocular pressure, damage to RGC axon bundles, optic nerve transection, and aging on mouse retinas.In terms of clinical applications, the article reviews research related to the use of Vis-OCT for retinal diseases such as glaucoma, retinal occlusive disease, diabetic retinopathy, and sickle cell retinopathy. At the same time, the article compares Vis-OCT with NIR-OCT in human retinal imaging, demonstrating the advantages and disadvantages of Vis-OCT.Conclusions and ProspectsVis-OCT technology can achieve submicron-level retinal imaging resolution, which significantly improves image quality. Moreover, it quantifies sO2 through spectral analysis and, combined with blood flow measurements, provides a new means of evaluating retinal circulation and oxygen metabolism. The match between visible light and the retinal sensing spectrum allows Vis-OCT to more directly reflect the relationship between the retinal structure and function.However, Vis-OCT also has some limitations. Its comfort level is relatively low, which limits its clinical application. Additionally, noise from super-continuum spectral sources reduces its signal-to-noise ratio. Furthermore, Vis-OCT has a limited imaging depth, which makes it challenging to observe deep tissues.Nevertheless, the future of Vis-OCT is promising. The imaging speed and quality can be improved by optimizing the system structures and image algorithms, which will provide ophthalmologists with more accurate diagnostic criteria. After improving the technology and expanding the applications, Vis-OCT is expected to play a significant role in screening and diagnosing retinal diseases as well as evaluating their treatment outcomes.In conclusion, Vis-OCT exhibits significant value in retinal disease research and clinical practice because of its unique advantages. The ongoing technological advancements should lead to further innovations and breakthroughs in ophthalmology.
ObjectiveIn surgical navigation system-assisted pedicle screw fixation, preoperative and intraoperative point clouds registration accuracy is crucial for positioning and navigation. When the patient's preoperative space is accurately registered to the actual surgical space, the surgical instrument can be guided to the patient's surgical site, and the planned surgical path can be accurately implemented during the operation. The preoperative point cloud is obtained by reconstructing the patients' preoperative CT, while a structural light scanner obtains the intraoperative point cloud during the operation. The acquisition methods of two-point clouds differ; hence, their densities and initial poses are quite different. Therefore, they are cross-source point clouds. Moreover, the scanned intraoperative point cloud is a small part of an entire spine since the exposed spine is very limited during an operation. Hence, the overlapping ratio of the preoperative and intraoperative point clouds is low. Existing registration algorithms are prone to fail or derive a low accuracy in preoperative and intraoperative point cloud registration. To solve these problems, this study proposes a preoperative and intraoperative point clouds registration algorithm with cross-source and a small overlapping ratio for pedicle screw fixation.MethodsThis study proposes a preoperative and intraoperative point clouds registration algorithm with cross-source and low overlapping ratio based on Farthest Point Sampling (FPS). The proposed algorithm includes coarse and fine registration. The coarse registration comprises three steps. Firstly, the voxel filter was used to down-sample the intraoperative point cloud to bring its density close to the preoperative point cloud. Secondly, the Fast Point Feature Histogram (FPFH) features of the intraoperative point cloud were extracted. The FPS was used to sample the preoperative point cloud, and then the preoperative point cloud was divided into several local regions by kd tree algorithm. These local regions formed the candidate set. Thirdly, the candidate set was traversed to calculate the FPFH features of each local region. The Sample Consensus Initial Alignment (SAC-IA) feature matching method was to realize feature matching and pose transformation estimation of intraoperative point cloud. The distance errors deduced by the SAC-IA method between the intraoperative point cloud and local point cloud were compared and the local region with the minimum distance error was selected as the optimal local region. The transformation of the optimal local region was the intraoperative and preoperative points clouds coarse registration's transformation. In fine registration, the Iterative Closest Point (ICP) algorithm was adopted to further align the intraoperative and the preoperative point cloud. It is performed based on the coarse registration result. The optimal local region is used as the target point cloud at this stage. Using the FPS and SAC-IC methods, an optimal local region sampled from the preoperative point cloud is obtained, enabling the point clouds can align under large original pose difference and low overlapping ratio conditions. In fine registration, the ICP algorithm was adopted to further align the intraoperative and the preoperative point clouds. The fine registration was performed based on the coarse registration results. The optimal local region was used as the target point cloud at this stage.Results and DiscussionsBased on the FPS method, the preoperative point cloud is divided into several local regions (Fig. 2). An optimal local neighborhood is derived to complete registration with the intraoperative point cloud. This study adopts nine pairs of preoperative and intraoperative point clouds for testing, with different overlapping ratios and initial poses (Table 1). The visualization results of the registration process using the proposed algorithm are shown in Fig. 3, including the initial pose of nine pairs of point clouds, the sampling results of FPS, and the coarse and fine registration results. To evaluate the performance of the proposed algorithm, two state-of-the-art registration algorithms, FPFH+ICP and SHOT+ICP, are adopted for comparison (Table 2). The proposed algorithm achieves the minimum coarse registration error, providing better alignment for ICP fine registration. A comparison of the final registration transformation matrix with the ground truth shows that the average rotation error is 0.406° and the translation error is 0.474 mm, which meets clinical requirements. Simultaneously, the registration time of the proposed algorithm is less than 2 min, which is adequate for an operation. In addition, the registration success rates of the three algorithms in the experiment are compared (Table 3). The successfully registered FPFH+ICP and SHOT+ICP algorithm samples are 6 out of 9. While for the proposed algorithm, it is 9 out of 9, demonstrating that the registration success rate increased from 66.67% to 100%.ConclusionsThis study proposes a point cloud registration algorithm with cross-source and a low overlapping ratio for pedicle screw fixation. Through the registration of preoperative and intraoperative point clouds of a lumbar vertebra with an overlapping ratio of less than 3%, the experimental results show that the proposed algorithm based on FPS can resolve the problems of density differences, large initial pose differences, and low overlapping ratios in the preoperative and intraoperative point clouds registration of pedicle screw fixation assisted by a surgical navigation system. High precision registration can be realized, improving the accuracy and safety of surgical navigation systems. The research only considers the rigid transformation of preoperative and intraoperative point clouds. Preoperative and intraoperative intervertebral motion will be considered in the future to make the proposed system algorithm more suitable for clinical practice.
ObjectiveTwo-photon microscopy (TPM) imaging has been widely used in many fields, such as in vivo tumor imaging, neuroimaging, and brain disease research. However, the small field-of-view (FOV) in two-photon imaging (typically within diameter of 1 mm) limits its further application. Although the FOV can be extended through adaptive optics technology, the complex optical paths, additional device costs, and cumbersome operating procedures limit its promotion. In this study, we propose the use of deep learning technology instead of adaptive optics technology to expand the FOV of two-photon imaging. The large FOV of TPM can be realized without additional hardware (such as a special objective lens or phase compensation device). In addition, a BN-free attention activation residual U-Net (nBRAnet) network framework is designed for this imaging method, which can efficiently correct aberrations without requiring wavefront detection.MethodsCommercially available objectives have a nominal imaging FOV that has been calibrated by the manufacturer. Within the nominal FOV, the objective lens exhibits negligible aberrations. However, the aberrations increase dramatically beyond the nominal FOV. Therefore, the imaging FOV of the objective lens is limited to its nominal FOV. In this study, we improved the imaging quality of the FOV outside the nominal region by combining adaptive optics (AO) and deep learning. Aberrant and AO-corrected images were collected outside the nominal FOV. Thus, we obtained a paired dataset consisting of AO-corrected and uncorrected images. A supervised neural network was trained using the aberrated images as the input and the AO-corrected images as the output. After training, the images collected from regions outside the nominal FOV could be fed directly to the network. Aberration-corrected images were produced, and the imaging system could be used without AO hardware.Results and DiscussionsThe experimental test results include the imaging results of samples such as fluorescent beads with diameter of 1 μm and Thy1-GFP and CX3CR1-GFP mouse brain slices, and the results of the corresponding network output. The high peak signal-to-noise ratio (PSNR) values of the test output and ground truth demonstrate the feasibility of extending the FOV to TPM imaging using deep learning. At the same time, the intensity contrast between the nBRAnet network output image and the ground truth on the horizontal line is compared in detail (Figs. 3, 4, and 5). The extended FOV of different samples is randomly selected for analysis, and a high degree of coincidence is observed in the intensity comparison. The experimental results show that after using the network, both the resolution and fluorescence intensity can be restored to a level where there is almost no aberration, which is close to the result after correcting using AO hardware. To demonstrate the advantages of the network framework designed in this study, the traditional U-Net structure and the very deep super-resolution (VDSR) model are used to compare with ours. When using the same training dataset to train different models, we find that the experimental results of the VDSR model contain a considerable amount of noise, whereas the experimental results of the U-Net network lose some details (Fig. 6). The high PSNR values clearly demonstrate the strength of our nBRAnet network framework (Table 3).ConclusionsThis study provides a novel method to effectively extend the FOV of TPM imaging by designing an nBRAnet network framework. In other words, deep learning was used to enhance the acquired image and expand the nominal FOV for commercial objectives. The experimental results show that images from extended FOVs can be restored to their AO-corrected versions using the trained network. That is, deep learning technology could be used instead of AO hardware technology to expand the FOV of commercially available objectives. This simplifies the operation and reduces the system cost. An extended FOV obtained using deep learning can be employed for cross-regional or whole-brain imaging.
SignificanceBlood flow is an important parameter for measuring vital signs, and hemodynamic parameters are functional indicators of the microcirculatory system of the skin, brain, heart, liver, kidneys, and other organs. Therefore, dynamic blood flow monitoring has important application value and significance in clinical and basic life science fields, such as clinical diagnosis, intraoperative guidance, drug research, disease mechanism research, and neuroscience. Laser speckle contrast imaging (LSCI) is a full-field optical imaging technique that uses the spatial and temporal statistical properties of laser scattering intensity to monitor the blood flow of tissues in vivo. It uses simple equipment, is non-invasive, and has a fast imaging speed and high spatial resolution. Additionally, it does not require the injection of a contrast agent and can perform continuous measures for a long time. Consequently, it is widely used to measure microcirculatory blood flow parameters such as the vessel diameter, blood flow velocity, blood perfusion, and blood density in tissues and organs. It also can help doctors locate the lesion precisely with clear and accurate blood flow data,and then analyze the corresponding functional response and pathological mechanisms, which has become one of the most important tools for the clinical diagnosis of fundus diseases, skin diseases, brain diseases, and so on. In addition, it is also an important tool for basic life science research in drugs, cardiovascular and cerebrovascular diseases, and brain cognitive and behavioral sciences. Consequently, in-depth research on novel LSCI techniques with high imaging quality is valuable and significant for improving the quality of medical care and promoting the development of basic research in life science.ProgressIn the past decades, many researchers have conducted extensive researches on how to improve the quality of LSCI and expand the scope of LSCI applications, and they have had positive progress. For example, a few research groups like Luo Qingming and Li Pengcheng at Huazhong University of Science and Technology, and Tong Shanbao at Shanghai Jiao Tong University have worked on portable LSCI systems, high signal-to-noise ratio LSCI, and high resolution LSCI, which have promoted the development of LSCI in China. Researchers abroad like Boas at Boston University, Zakharov at the University of Fribourg, and Dunn at the University of Texas at Austin have worked on high-precision imaging using LSCI techniques, such as static scattered light correction and quantitative analysis of LSCI, which has also greatly promoted the development of key techniques and novel LSCI applications.In this paper, we presented a systematic, comprehensive and integrated analysis, review and summary of the current researches about key techniques and applications of novel LSCI at home and abroad emphatically from the aspect of high signal-to-noise ratio LSCI, high-resolution LSCI, high-precision LSCI, large imaging depth LSCI, and novel LSCI systems based on the investigations of current literature. In this way, we can help researchers learn more about the frontier technologies of LSCI and understand the technical challenges we faced, and we can provide ideas with a reference value to promote the development of high-quality, highly practical, and innovative LSCI systems to meet the needs of clinical diagnosis and basic biomedical research. The review consisted of the following contents: First, the technical problems of measuring deep blood flow and achieving high resolution, high signal-to-noise ratio, and high precision have been systematically summarized, and the corresponding solutions are indicated. Subsequently, we review high signal-to-noise ratio LSCI techniques based on anisotropic filtering, eigenvalue-decomposition, and transformation domain collaborative filtering methods. Meanwhile, high-resolution LSCI for motion artifact, out-of-focus blur, and non-uniform light intensity correction are also summarized. Third, we elaborate the high-precision LSCI from the perspective of static scattered light correction, quantitative analysis, and novel LSCI algorithms. After summarizing the LSCI with a large imaging depth, we introduce the latest research on the novel LSCI system and its applications in the fields of cortical blood flow imaging, surgical and therapeutic procedures, and brain and cognitive-behavioral sciences. Finally, we discuss and look forward to the development of LSCI in the future.Conclusions and ProspectsIn conclusion, LSCI has made qualitative leaps and developments in theory, imaging systems, computational methods, and clinical applications. The imaging quality of LSCI has been developed to have a high signal-to-noise ratio, high resolution, high accuracy, and large imaging depth. However, as the application scenarios of LSCI become more and more complex, which introduces greater challenges to the development of key techniques and application of LSCI. In the future, LSCI will be deeply integrated with emerging interdisciplinary fields such as biomedicine, optoelectronic information, artificial intelligence, and big data. In addition, new breakthroughs are expected in the following respects. (1) Quantitative analysis capacity. The capacity is still an important fundamental issue for LSCI in functional applications. (2) Combination of LSCI with new endoscopic technology (this will enable the noninvasive measurement of blood flow). (3) Miniaturization and integration. The development of new materials and electronic devices will certainly promote the miniaturization and integration of new LSCI systems. (4) Combination of LSCI with artificial intelligence. Artificial intelligence will further promote the development of LSCI technologies and their applications. (5) Combination with other imaging modalities (this will build a new model for LSCI-based multimodal clinical diagnostic applications). It is believed that LSCI will show a synergistic development trend in the future. We look forward to seeing the development of technologies and applications of LSCI.
SignificanceRetinal opto-physiology is the physiological response of the retina to a visible light stimulus, reflecting the function of the retina to a certain extent. Optoretinography, also termed optoretinogram (ORG), is a newly developed functional imaging technique for precisely quantifying the opto-physiological response of the retina. It uses optical coherence tomography combined with controllable light stimulus, to accurately measure retinal morphology and optical property changes in response to a light stimulus by detecting the peak position and scattering intensity alternations in optical coherence tomography (OCT) images. Moreover, ORG can achieve microscale lateral resolution, nanoscale axial resolution, and millisecond temporal resolution by combining adaptive optics and phase analysis techniques, and be used to measure opto-physiological functions of the retina at the cellular level. To perform ORG, only an optical stimulus unit is required to be added to the existing OCT system. Currently, it is still in the stage of technology development and mechanism exploration. Once standards are established, ORG may be used in broad ophthalmic research and clinical practice. In this study, the history of OCT functional imaging development for probing the retinal opto-physiological signal is systematically reviewed, the latest progress in ORG technology is summarized, and several future directions of ORG technology are discussed.ProgressBecause of the excellent spatial resolution of OCT and its extensive use in basic research, clinical diagnosis, and treatment, researchers have been committed to capturing the functional response of nerve cells to a visible light stimulus using OCT. Early studies of OCT photo-physiological functional imaging focused on finding the changes in retinal scattering signals after light stimulation. With the gradual improvement of OCT performance (such as resolution and imaging speed) and the wide application of in vivo imaging technology, a series of breakthroughs have been made in the field of OCT retinal functional imaging in recent years. The optical path difference changes of cone outer segment in living human retina after a light stimulus [Fig. 2(a)-(c)] were successfully measured using high-speed full-field OCT combined with phase analysis technology. Subsequently, Zhang et al. used OCT to observe the function signals of mouse retina in response to the light stimulation with different intensities. They reported the changes in the thickness of the rod outer segment at a micron level and changes in scattering signal intensities of several retinal layers [Fig. 2(d)-(h)]. Moreover, they confirmed that the response signals came from the visual photo-transduction process using gene knockout mice. Furthermore, due to the advantages of adaptive optics enhanced OCT (AO-OCT) in imaging resolution, researchers have successfully measured the photo-physiological signal at the level of a single cone cell in the human eye. For example, Zhang et al. measured the functional response of human cone cells after light stimulation using AO-OCT in 2019 and successfully distinguished three types of cone cells in the human eyes via different cone cell responses to different color stimuli (Fig. 3). In the recent studies, several experimental groups have conducted the diseases or mechanism research on the retina from a physiological perspective. Qian et al. changed the mouse retina through transgenic or drug methods to carry out controlled experiments with conventional wild mice. They found that changes in the thickness of the mouse’s external retina after light stimulation were affected by the base level of mitochondrial respiration and oxidative stress reaction. This suggests the favorable conditions for the clinical application of OCT-based photo-physiological functional imaging.Conclusions and ProspectsThe method based on OCT retinal opto-physiological functional imaging and visible light stimulation is collectively called optoretinogram. ORG is a new technology in which the obtained opto-physiological signals can reflect the function of retinal tissue. This addresses the limitation of conventional OCT imaging technology which can only provide details of the retinal structure. Moreover, studies of human and animal retina indicate that retinal responses to light stimulation involve subtle changes in several structures including the rod, cone, retinal pigment epithelium, Bruch's membrane, and choroid. All are closely related to the structures affected by various retinal diseases. Thus, to some extent, early retinopathy may affect the intensity and change rate of retinal opto-physiological function signal. Therefore, this can provide a new diagnostic basis for detection of early disease by measuring the abnormal changes in the opto-physiological function signal.Future developments of ORG may include the following four aspects: 1) realizing the local analytical capability of ORG while maintaining wide-field macroscopic imaging; 2) further enhancement of the sensitivity of OCT to enable detection of weaker functional signals; 3) extracting the functional signal quickly and automatically and exploring the characteristic functional signal of early retinopathy; and 4) optimization and standardization of experimental methods.
ObjectiveThe Muller matrix, as a method for characterizing the polarization properties of samples, contains complete information about the polarization properties of samples, and it has become an important indicator for characterizing pathological tissues in basic and preclinical studies. However, in the traditional polarized light imaging method for measuring the Muller matrix, the scattering depth of polarized light in collagen tissue cannot be controlled. The obtained Muller matrix information is the average of unknown depths in collagen tissue, and it is impossible to accurately measure the Muller matrix information of the pathological tissue area. Polarized spatial frequency domain imaging (PSFDI), which combines spatial frequency domain imaging (SFDI) and polarized light imaging, is applied to measure the optical properties of biological tissues accurately.MethodsSFDI relates the spatial frequency of the projected stripe pattern to the penetration depth of the detected light, and the imaging depth can be controlled by controlling the spatial frequency of the projected light. We designed and validated a polarization SFDI system that uses the SFDI technique to control the imaging depth, projects the streak pattern onto the surface of the measured tissue, constructs a polarizer and detector to modulate the polarization state of the polarized light, and then acquires the image data using a CMOS camera and calculates the Mueller matrix information.Results and DiscussionsExperimental results showed that the grey-scale plate diffuse reflectance measured by the polarization SFDI system was linearly correlated with the standard value (R2=0.99988). The depolarization coefficient tends to be proportional to the fat emulsion volume fraction, the two-way attenuation coefficient increases with the increase of two-way attenuation owing to the two-way attenuator, and the accurate measurement of the phase delay of the quarter-wave and full-wave plates indicates that the system can accurately measure the sample polarization parameters. A comparison of uniform light field illumination and polarization-sensitive SFDI shows that the latter effectively controls the depth and accurately measures the shallow Mueller matrix of the sample. The results of this study are expected to effectively improve the accuracy of the detection of polarization characteristics of superficial tissues and promote early tumor detection.ConclusionsIn this study, a PSFDI system is developed based on polarized light imaging and SFDI, and the device structure, measurement method, and data processing method are introduced. By performing the error calibration of the PSFDI device, the measurement error of the device can be less than 2%. By performing Mueller matrix imaging on tissues, we verified the reliability of the device to measure tissue polarization Mueller and the accuracy of the Mueller matrix decomposition; hence, PSFDI can be used to obtain the optical properties of various samples. The PSFDI can accurately image pathological regions, providing accurate physiological parameters for pathological analysis, and it has a wide range of biomedical applications.
ObjectiveMicrocirculatory dysfunction may cause circulatory failure, insufficient oxygen delivery, and fatal risks. Microscopes are used to observe microcirculation, but they can only image superficial tissues. In addition, they can hardly provide functional information. In this study, we report a photoacoustic endoscope for in vivo imaging of the gastrointestinal microcirculation. The imaging probe is inserted into the rectum of a small animal for rotational-scanning endoscopic imaging. The vascular structures in the gastrointestinal wall can be visualized by detecting the ultrasound excited by the pulsed laser. Moreover, the blood oxygen saturation can be measured and imaged with a dual-wavelength excitation based on the difference in the optical absorption spectrum between oxy- and deoxygenated hemoglobin. We believe that this technology is capable of detecting the functional changes associated with microcirculation diseases with minimal invasion.MethodsThe imaging system consists of an endoscopic imaging probe, dual-wavelength pulsed laser source, rotary scanning device, and data acquisition and control module. First, we design an all-fiber endoscope probe containing two functional optical fibers as follows: one responsible for guiding and focusing the pulsed light and the other equipped with a laser ultrasonic sensor to detect the photoacoustic signal. Second, we design a rotational scanning device that rotates synchronously with the probe to achieve fast and unidirectional rotary scanning. This is achieved by miniaturizing the 980-nm pump laser, the optical amplifier, and the photodetector. Finally, we perform high-resolution in vivo endoscopic imaging of the rat rectum.Results and DiscussionsThe endoscope probe has a diameter of 2.75 mm, a resolution of 12.5 μm, signal jitter root mean square of 2.5%, and a B-scan frequency of 1 Hz. The instrument is stable and provides spatial resolution in high-speed scanning and is suitable for small animal digestive tract endoscopic imaging. The functional imaging results of the rectum of healthy rats show that we achieve 360°scanning, obtain the three-dimensional imaging results of hemoglobin concentration distribution, and show the vascular structure of the inner wall of the rat rectum. Along with the spatial distribution of blood oxygen saturation, the images show the distributions of the artery and vein in the inner wall (Fig. 3). The imaging results of septic rats show the changes in microcirculation. According to the imaging result, the number of blood vessels in the intestine of rats gradually decreases, and the blood oxygen saturation also declines in 5 h (Fig. 4). The above results reflect the phenomenon of insufficient tissue perfusion caused by sepsis.ConclusionsIn summary, we develop a photoacoustic endoscope for in vivo rectal imaging. By using fiber optic ultrasound sensors, the endoscope can image the vascular structure and visualize the changes in oxygen saturation. By using this endoscope, we can visualize the gastrointestinal microcirculatory disorder caused by lesions. The structure, number, and blood oxygen saturation level of blood vessels are significantly changed. The experimental results show that this technology can provide functional imaging results with high spatial resolution and high contrast in the endoscopic imaging of narrow cavity structures, thus providing a feasible imaging method for the characterization of microcirculation status and the diagnosis and treatment of acute and severe diseases.
ObjectiveLipid droplets are important organelles closely associated with various cellular physiological activities. Confocal fluorescence imaging is a powerful tool for observing lipid droplets and studying their diverse functions. However, lipid droplet fluorescent probes with the high fluorescence intensity and labeling selectivity required for cellular lipid droplet fluorescence imaging are limited, severely limiting the in-depth study of lipid droplets. In this study, we develop Lipi-QL, a quinoline-derivative lipid droplet fluorescent probe with fluorescence-switching properties.MethodsThe probe exhibits high selectivity for lipid droplet labeling owing to its sensitive polar quenching fluorescence properties. The donor-type molecular structure also confers high fluorescence intensity and large Stokes shifts on the probe. When using this probe for confocal fluorescence imaging of cellular lipid droplets, significantly better labeling selectivity is achieved at varying concentrations than when using the commercial BODIPY 493/503 lipid droplet probe. Additionally, three-dimensional confocal imaging of fixed cells and four-color confocal imaging of live cells are performed using this fluorescent probe. The development of this probe provides a powerful tool for studying the physiological functions of lipid droplets and provides a new idea for the design of new highly labeled selective fluorescent probes.Results and DiscussionsAs shown in Fig.1(c), the probe exhibits highly efficient fluorescence emission when the water volume fraction is 0, indicating that it can exhibit high fluorescence intensity within lipid droplets. When the water volume fraction gradually increases, the probe exhibits extremely sensitive fluorescence quenching properties: quenching most of the fluorescence emission when the water volume fraction is only 1%. When the water volume fraction increases to 20%, the probe's emission is almost completely quenched, and the fluorescence signal disappears. This indicates that even if a small portion of the probe enters the cell and stains organelles other than lipid droplets, the fluorescence emission is quenched by the polar environment in which it is placed, thus showing a high selectivity for lipid droplet staining. We also test the fluorescence switching characteristics of the commercial lipid droplet dye, BODIPY 493/503. As shown in Fig.1(d), the fluorescence quenching of BODIPY 493/503 in the dioxane solution with 40% water volume fraction is not apparent, which may be the main reason for its poor lipid droplet staining selectivity. Figure 3 shows that the Lipi-QL fluorescent probe efficiently stains cellular lipid droplets at different concentrations. In contrast, BODIPY 493/503 stains lipid droplets much less selectively, staining other membrane-like cellular structures in addition to cellular lipid droplets with a lower imaging signal-to-noise ratio. This staining selectivity comparison highlights the significant advantage of the polar quenching luminescence property of the Lipi-QL fluorescent probe for the efficient and selective labeling of cellular lipid droplets. After washing the free probe with phosphate buffered saline (PBS), three-dimensional confocal imaging is performed. The experiment is performed at a high xy-plane point resolution with a small z-sweep step (200 nm) to obtain high-quality 3D confocal photographs (Fig.4). The spatial distribution of intracellular lipid droplets can be seen clearly in this photograph, demonstrating the usefulness of the probe for 3D confocal imaging. The Lipi-QL fluorescent probe is also used for multicolor confocal imaging because of its excellent performance. The nuclei, lipid droplets, lysosomes, and mitochondria of live HeLa cells are stained with the Hoechst 33342 commercial dye for nuclei, Lipi-QL commercial dye for lipid droplets, LysoTracker Deep Red commercial dye for lysosomes, and MitoTracker Deep Red commercial dye for mitochondria, respectively. High-quality four-color confocal images of living cells are successfully obtained by performing confocal fluorescence. Based on the different absorption and emission spectra of these four fluorescent probes, imaging is performed through line-by-line scanning, effectively avoiding the occurrence of crosstalk between individual fluorescent channels.ConclusionsIn conclusion, an advanced lipid droplet fluorescent probe with fluorescence switching properties, Lipi-QL, is developed in this study, which allows for the efficient and selective labeling of cellular lipid droplets. The probe also has high fluorescence brightness, a large Stokes shift, and good biocompatibility. Based on these excellent properties, high-quality three-dimensional confocal imaging of fixed cells and four-color confocal imaging of live cells are successfully achieved using this probe, highlighting its utility in lipid droplet fluorescence imaging. The development of this probe provides an effective tool for cell biology studies of lipid droplets and a new approach for the design and synthesis of highly labeled selective fluorescent probes.
ObjectiveImage-based non-contact measurements for pulse wave remote acquisition and monitoring have an important practical value in clinical use. Accurate pulse waves are a major prerequisite for measuring parameters of human physiology such as the heart rate, heart rate variability, blood oxygen concentration, and blood pressure. Based on the fact that the carotid artery is the closest observable artery to the human heart and contains a wealth of physiological information, vibrations of the epidermis caused by blood flow can be observed on the surface of the human body. In addition, the amplitude of random motion on the neck is much smaller than that on the human face. Accordingly, the signal source is set on the neck for better observations, less disturbance, and more up-to-date results. Under normal circumstances, the pulsation of the human carotid artery causes a small vibration that is visible to the naked eye and can be obtained by analyzing the vibration using conventional image and signal processing methods. However, in clinical practice, some patients have a relatively weak carotid pulse, and the existing statistical signal processing and time-frequency domain signal processing methods are inadequate for obtaining the desired signal. Thus, a new signal processing method is required for these types of situations.MethodUnder the illumination of an 850 nm near-infrared light source, a near-infrared camera was used to continuously shoot the image sequence of the vibration of the neck skin. The final signal was obtained through a series of images and signal processing. The specific process is described as follows. First, the region of interest (ROI) was obtained using the inter-frame difference method. The original gray signal was then obtained by calculating the mean value of the ROI. Next, the original signal was normalized from the gray signal at an interval of 0 to 1. Finally, the desired pulse wave signal was acquired using bandpass filtering and the proposed multi-region dominant frequency enhancement (MRDFE) method. The MRDFE method is a joint algorithm that combines frequency domain processing and principal component analysis in two steps. In the first step, the signal obtained in each ROI was assigned the weight of the dominant frequency signal-to-noise ratio. In the second step, the signals in these ROI channels were evaluated by principal component analysis, and the feature vector corresponding to the first eigenvalue obtained was the final output signal. To further demonstrate the robustness of the algorithm, we established our own database, which contained 24 sets of weak pulse vibration image sequences. In dealing with these data, we compared our method with other existing algorithms based on four indicators: periodic integrity, periodic variation, tidal wave integrity, and repulse wave integrity.Results and DiscussionsThe proposed MRDFE method can be used to obtain pulse waves with preserved feature points in a weak pulse situation (Fig. 4). To compare the MRDFE method with other conventional methods, a feature point recognition algorithm called the stepwise threshold descent method was used to detect feature points from the final signal obtained by each method. Our experimental results show that the proposed method performs much better than the other three conventional algorithms. Our method exhibits a more stable periodic state and retains approximately 70% of the tidal wave characteristics and more than 50% of the repulse wave characteristics (Table 1). Based on observations of the signals derived from the different methods (Fig. 7), the periodicity of the pulse wave obtained by our method is more obvious, and more feature points are preserved. The MRDFE method enhances the signal with a high signal-to-noise ratio and weakens the signal with a low signal-to-noise ratio through weight assignment, yielding satisfactory results.ConclusionsThis study presents a method for obtaining pulse wave signals under the condition of weak pulse vibrations of the carotid artery. With a near-infrared light source used for illumination, the image sequence of neck skin vibration was captured by a camera. Several ROIs were selected from the image sequence, and the initial signal was acquired using outlier processing and bandpass filtering. The pulse wave signal of the weak pulse vibration was then processed successfully using the MRDFE method. Compared with other signal processing methods, the analytical results show that the signal obtained by the MRDFE method is of higher quality, preserves a greater number of feature points, and provides better cycle integrity. Our analysis and experimental results show that the proposed method is superior in performance to the existing signal processing methods. Robust and reliable pulse wave signals can be obtained using this method and applied in further measurements of the heart rate, heart rate variability, blood oxygen, and even blood pressure. The MRDFE method adds considerable value to new signal processing for image-based non-contact pulse wave extraction.
ObjectiveIn typical tumor therapy, the drug must reach the tumor site via blood vessels and access the cell membrane of the tumor cells to act on a certain target. The drug recognizes the target molecules and then enters the tumor cells in a specific manner that facilitates the release of the drug without toxic side effects on normal cells. Numerous membrane proteins and receptors on the cell membrane can be considered targets during drug carrier design. Corresponding biochemical studies, such as immunoblotting and flow cytometry, are often conducted, supplemented by fluorescence co-staining imaging. However, intensity-based fluorescence imaging has considerable limitations, both in terms of its inability to distinguish small-molecule drugs from nanomedicine drugs and to monitor endocytosis in living cells in real time. Fluorescence lifetime imaging microscopy (FLIM) is commonly used to evaluate the lifetime of fluorescent moieties in living cells for quantitative microscopic analysis. Förster resonance energy transfer (FRET) can be used to characterize the transfer of energy from a donor fluorescent molecule to an acceptor fluorescent molecule. FLIM technology with FRET (FLIM-FRET) can monitor protein interactions and the dynamic processes of subcellular organelles in living cells.MethodsIn this study, doxorubicin (DOX) nanoparticles encapsulated in bovine serum albumin (BSA) were synthesized. Albumin nanoparticles demonstrate good biocompatibility and inherent passive targeting in living organisms and can be effective drug carriers for slow release and reduced toxic side effects. DOX is an amphiphilic molecule that is not completely encapsulated in the nanoparticles and is attached to the nanoparticle surface. Superfolder GFP (sfGFP) was transfected into the cell membrane as a donor, and the BSA-DOX nanoparticles were used as acceptor molecules. Together, both molecules constituted the FRET nanosystem. During the uptake of nano drugs by cells via endocytosis, the distance between the cell membrane and nano drug meets the criteria for FRET induction, and the fluorescence lifetime of the donor is shortened during the process. When the endocytic vesicles release the drug intracellularly, the distance between the cell membrane and nano drug is altered, and the FRET effect diminishes or disappears. In this study, we used a two-photon excitation fluorescence lifetime microscopic imaging system (TP-FLIM) to monitor the FRET effect during this process, to distinguish between the diffusion movement of nanoparticles being endocytosed into cells and small-molecule drugs and to monitor the endocytosis process of cells in real time. We used this method to verify the upregulation of the endocytosis movement of cells under starvation conditions.Results and DiscussionsIn this study, BSA was used to wrap DOX into nanoparticles that could be endocytosed into cells, resulting in the formation of BSA-DOX nanoparticles with a particle size below 100 nm. The process of cellular uptake of nanoparticles by endocytosis is long, which enables a more in-depth study of microscopic physiological processes. In addition, the endocytosis pathway of the nanocarriers was evaluated using four endocytosis pathway inhibitors. BSA-DOX nanoparticles entered the cells via clathrin-mediated endocytosis. The associated dynamic process was elucidated. Our study shows that the FLIM-FRET technique combined with quantitative analysis methods can be used to study the similarities and differences between small-molecule drugs and nanoparticle-cell interactions.ConclusionsIn this study, we present a new method for the qualitative and quantitative analysis of endocytosis of nanomedicine in OVCAR-3 cells. We synthesized BSA-DOX nanoparticles by desolvation using a material that allows us to use its own fluorescence to form FRET pairs with sfGFP proteins transfected onto the cell membrane. We used the TP-FLIM system for qualitative analysis of cellular endocytosis by two-photon fluorescence and interference-free monitoring of donor lifetime during FRET. Quantitative analysis was performed by FLIM. The experimental results show that the distance between the cell membrane and nano molecule can be accurately reflected by detecting FRET efficiency as the nanomedicine is endocytosed by the cells and released within the cells. We also used this method to verify that starvation-treated cells upregulated endocytosis motility.
ObjectiveExpansion super-resolution technology, in which resolution is improved by improving the corresponding sample, has emerged in recent years. Owing to its strong compatibility with other optical technologies and its high resolution, it has attracted an increasing amount of research attention. The combination of expansion super-resolution technology and other super-resolution techniques is one main development direction for expansion super-resolution technology. Expansion combined with optical fluctuation super-resolution technology (ExM-SOFI) is a widely used compound expansion technology with relatively few restrictions. To enhance the imaging of the existing ExM-SOFI, we applied an imaging buffer to enhance the anti-quenching ability of the expansion sample during shooting. The fluorescence intensity, fluorescence fluctuation amplitude, and on-time ratio of common dyes in ExM-SOFI were improved. Finally, the staining results of the microtubules and vesicles indicate that the use of this technique can make the sample more realistic, with fewer artifacts, and can improve the final resolution of expansion samples in high-order SOFI technology.MethodsIn this study, we derived an anti-quenching-enhanced ExM-SOFI technique by improving the existing ExM-SOFI technique using an imaging buffer. First, the samples were labeled with biotinylated antibodies. Biotins can retain recognition sites after expansion, for post-expansion staining, to reduce signal loss. An expanded hydrogel was then obtained using a common expansion protocol. Next, the expanded hydrogel was cut into a suitable size and re-embedded in a high-concentration solution to prevent it from shrinking. After re-embedding, the expanded hydrogel was incubated with a dye modified with streptavidin. During photography, the stained hydrogel was immersed in an imaging buffer for imaging. We used an imaging buffer with an oxygen-scavenging system as the main component. The fluorescence intensity, anti-quenching ability, and fluorescence fluctuation amplitude of the images before and after the buffer treatment were analyzed. In addition, the on-time ratio and artifacts of the SOFI images before and after buffer treatment were analyzed, and the changes in different orders of SOFI were compared.Results and DiscussionsThe experimental design is illustrated in Fig. 1. According to the analysis results of the images before and after the imaging buffer treatment, the fluorescence intensity of the sample with the imaging buffer was approximately 60% higher than that without the imaging buffer [Fig. 2(c)]. The signal quenching speed of the sample with the imaging buffer was slower during the shooting process compared with that of the sample without the imaging buffer [Fig. 2(d)]. In the analysis of the fluorescence fluctuation amplitude [Fig. 2(e)], the fluorescence fluctuation amplitude of the image after the buffer was added was several times larger than that of the image before the buffer was added. Enhancements in the fluorescence intensity, anti-quenching ability, and fluorescence fluctuation amplitude are important to improve the quality and resolution of SOFI imaging. The on-time ratio is an important parameter that affects the imaging quality of SOFI; conventional dyes are often not conducive to SOFI analysis because of their high on-time ratios. We analyzed the on-time ratio of the images before and after the imaging buffer was added, and the results are shown in Fig. 3. Compared with the image without a buffer, the overall on-time ratio of the image with a buffer decreased from 80%-95% to 35%-40%. In a study by Wang et al. on SOFI, an on-time ratio in this interval was better for SOFI analysis. In addition, we analyzed the resolution-scaled Pearson (RSP) correlation values before and after the buffer was added; higher values indicate a better agreement. The RSP value after the buffer was added was higher than that before, indicating that the image with buffer was more authentic. In the comparison of different orders of SOFI (Figs. 4 and 5), in the imaging results of both the microtubule or vesicle, fewer image artifacts existed after the buffer treatment than before, and the real structure was better maintained in high-order SOFI.ConclusionsExM-SOFI technology is a composite expansion technology that has relatively few equipment limitations and can increase resolution. However, owing to the loss and dilution of fluorescence signals during the preparation of expanded samples, the signal of the expanded samples can be weak, making it difficult to achieve the best results during SOFI continuous shooting. In this study, we proposed an anti-quenching enhanced ExM-SOFI technology that combines imaging buffer technology with the original ExM-SOFI to reduce fluorescence quenching during shooting. We found that this technique enhanced both the intensity and fluctuation amplitude of the fluorescence. The on-time ratio was also reduced to a range that was more suitable for SOFI analysis, enabling ordinary dyes to perform better in ExM-SOFI. An increase in the RSP value also indicated that this technique increases the credibility of the image. Finally, a comparison of different order SOFI images showed that this method reduced artifacts and better maintained the real structure in high-order SOFI.
ObjectiveThe physical structure, physiological characteristics, and health status of skin tissues are closely related to the quality of daily life and safety of the human body. Measuring human facial skin structures and characteristics has attracted increasing attention recently, because it can reveal the health conditions of the body and the dense distribution and complex structures of skin tissues in different regions. Polarization imaging is a non-contact label-free method that can provide abundant optical and structural information of tissues. Thus, it has been applied on many biomedical studies including facial skin tissue detection. However, there is still a lack of systematic and quantitative studies on the skin tissue characterization of different facial regions using polarization imaging parameters, which hinders further dermatological applications of polarimetry. To deal with this problem, we proposed a non-contact and in vivo measurement method of facial skin structures and characteristics based on polarization imaging; this method can obtain the heterogeneous distribution of five skin parameters in different facial regions under three polarization modes. Our results suggested ways of improving the effectiveness of non-contact and quantitative measurements of appropriate facial skin parameters by polarization imaging in future dermatological applications.MethodsFor human facial skin imaging, we first developed a polarization-adjustable instrument that is mainly composed of a light source, the linear polarization modulators, and a camera. It can extract the information of the skin surface and deep skin layer by using three polarization imaging modes, namely, non-polarized imaging (NPI), parallel-polarized imaging (PPI), and cross-polarized imaging (CPI). We recruited 15 healthy male (age 30.4±7.7 years old) and 5 female (age 29.4±7.4 years old) volunteers, respectively, their front, left, and right facial regions were imaged under the three polarization modes introduced above. The L*, b*, a*, and individual typological angle (ITA) parameters based on the CIE Lab color space, which are often used to assist dermatological studies, and the t parameter of the skin texture feature extracted by the Frangi filter were selected to further analyze the measurement results of distinct facial regions (cheek, upper eyelid, forehead, nose, and perioral area). The skin structures and characteristics of different facial anatomical regions of the volunteers were evaluated by dermatologists of the China Academy of Chinese Medical Sciences as the analysis criteria in this study.Results and DiscussionsThe results indicated that there are significant differences in the measurement results of five parameters for the subjects’ whole facial skin under different polarization imaging modes (PPL* and ITA are related to skin pigmentation, which can hardly be evaluated by polarization imaging modes (Fig. 3 and Fig. 6). The parameters a* and b* focus on analyzing the degree of deposition information of chromophores in the deeper skin layers, which can be successfully evaluated by the CPI mode (Figs. 4 and 5). Specifically, among the selected facial regions, the cheek region has the lowest combined concentration of chromophores in the epidermis and dermis, showing the lightest skin tone. Similarly, the forehead region is the most hyperpigmented. For the parameters of skin texture, the t parameter is related to skin aging and hydration. Compared with the NPI mode, the PPI mode can better reflect the length, width, and depth of skin stratum corneum wrinkles and hair shaft information, while the t parameter in the CPI mode can provide hair shaft texture information (Fig. 7).ConclusionsIn this study, we proposed a non-contact and in vivo measurement method of human facial skin structures and characteristics based on polarization imaging. The influence of different polarization imaging modes on the measurement of five facial skin parameters is analyzed. Using this imaging system, we quantitatively explored the effect of three different polarization imaging modes, namely, NPI, PPI, and CPI, on the measurement of facial skin parameters of L*, a*, b*, ITA, and t. The results showed that for certain facial skin parameters, the selection of an appropriate polarization imaging mode for different facial anatomical regions can improve the measurement accuracy, better characterize the facial skin tissue structure, composition, and metabolism of the region of interest, and facilitate the evaluation of facial skin health conditions and detection of pathologies. Simultaneously, the results suggested that for the quantitative detection of other body skin areas, we still need to establish a clear relationship between the corresponding skin structure, composition, metabolism, and polarization imaging parameters.
ObjectiveOptical diffractive tomographic microscopy is a new wide-field, non-invasive and label-free three-dimensional (3D) imaging technology for cells and tissues, which has great application prospects in cell metabolism, pathology and tumor diagnosis. However, with the continuous development of modern biological research, the field-of-view (FOV) of traditional optical diffractive tomography (ODT) cannot meet the needs of observation any more. The invention of large field-of-view ODT technology, while maintaining subcellular resolution, is in an increasingly urgent need.At present, various quantitative phase imaging technologies require higher spatial-bandwidth product. For example, the sampling rate of interference streaks acquisition in off-axis holographic imaging is more than three times that of intensity imaging. In the condition of a certain number of camera pixels, only a small FOV can be acquired. In order to conduct large-FOV quantitative phase imaging, the number of pixels in the single image is doubled, and the data flux of images is too large. It leads to the facts that the image storage becomes more difficult, the complexity of recovery algorithm is aggravated, and the time of settlement increases.MethodsThe traditional method to realize large FOV is to scan different areas and then splice the images. However, the method is not suitable for living cells since they constantly move, which limits the further application of the traditional ODT method in biology. To solve the problem, we propose a new ODT technology which can realize the large FOV.Based on the Mach-Zehnder transmission holographic imaging system, we make some unique designs for large-FOV imaging requirements. The main innovations are described here.Firstly, we design a non-destructive pupil holographic beam binding scheme. We use the D-shaped mirror instead of beam splitters for beam combining. It can achieve zero loss of intensity and unlimited size. Secondly, we achieve large-FOV oblique plane illumination under the large numerical aperture (NA). Finally, we improve the image acquisition system. We choose a 21 megapixels camera and the faster CoaXPress-12 card as the data acquisition card, and achieve the 50 Gbit/s data flux. The optical elements and galvanometer of the system are re-selected to ensure that there is no aperture limitation.In addition, we rewrite the data processing program, considering the large amount of data in the large-FOV ODT system. We reconstruct the software of images acquisition to realize the high-speed image acquisition and storage. Then, we edit the new multithread ODT recovery algorithm based on C++ for 64-bit system, which can automatically recover all the collected data synchronously. Using the above system and algorithm, we image 5 μm polystyrene microspheres to verify the feasibility of the system. Then, Hela cells are imaged, which verifies that the method has long-term 3D observation ability for dense tissue cells and living cells.Results and DiscussionsIn this paper, a large-FOV optical diffraction tomography technique is proposed. The large-FOV ODT uses all the FOV of the objective lens to reach the limit of the imaging range. At the same time, it has both high resolution and long-term 3D imaging capability for living cells. Compared with the traditional ODT system, the imaging range of the proposed system is larger [Figs. 3(a) and 5(a)]; more photons scattered by complex samples can be obtained, so that the signal-to-noise ratio (SNR) is better [Figs. 3(b) and 3(c)]. Moreover, the ringing and artifacts effects of the edge are smaller [Figs. 3(b), 3(c), 5(b), and 5(c)]. The interaction between cells, as well as more cells in different states, can be observed simultaneously in a FOV (Figs. 5 and 6).ConclusionsThe results show that the large-FOV optical diffraction tomography technology has both subcellular resolution and long-term 3D observation ability of label-free living cells. Compared with the traditional system, the large-FOV ODT system has smaller edge effects and obtains more information of cells, so it is beneficial to observing the interaction between cells, and is helpful to realizing the long-term 3D observation of huge living cells such as oocytes. It will have more biological applications.
ObjectiveHeart valve disease is a growing public health concern worldwide. A prosthetic heart valve is a heart implant intervention medical device for the treatment of heart valve disease, which mainly includes bioprosthetic and mechanical valves. Diseased native valves are often replaced with bioprosthetic valves made from porcine or bovine pericardium, which has a lower risk of thrombosis and hemodynamic advantages than mechanical valves. Nonetheless, bioprosthetic valves do not have long-term durability, mainly because of their early structural failure. Therefore, an in vitro fatigue test is required for manufactured bioprosthetic valves; further, it is very important to evaluate the quality of the valve after the fatigue test, thereby obtaining a basis for the optimization of valve performance.MethodsThe OCT light source was a MEMS-tunable vertical cavity surface-emitting laser (VCSEL, Thorlabs, SL131090). The laser could sweep at a rate of 100 kHz over a broad spectral bandwidth of ~100 nm with a center wavelength of 1300 nm, providing an experimental axial resolution of ~16 μm and an imaging depth of ~11 mm in air. The output light from the laser source was first fiber-coupled into an interferometer, where the light was split by an 80:20 fiber coupler into a sample arm and reference arm. In the OCT sample arm, a scanning lens (Thorlabs, LSM05) with an effective focal length of 54 mm was used to collimate the detection light on the sample, providing an experimental lateral resolution of ~32 μm, and an X-Y galvanometer was adopted for three-dimensional (3D) volume scanning. The light backscattered from the sample was recombined with the light reflected from the reference mirror, and the interference signal was detected using a balanced detector (Thorlabs, PDB470C). A stepwise raster scanning protocol (Z-X-Y) was used for volumetric imaging, with 1000 A-lines per B-frame (fast-scan, X-direction) and 1000 B-frames at 1000 tomographic positions per volume (slow-scan, Y-direction). OCT imaging covered a field of view (FOV) of 12 mm (X)×12 mm (Y) of the swine heart valve leaflets, and a wide FOV of 28.5 mm (X)×28.5 mm (Y) of the bioprosthetic valves. The captured interference data were converted to amplitude form using a fast Fourier transformation (FFT) processed on the MATLAB (MathWorks) platform. The bioprosthetic valve surface boundary fitting algorithm transforms the depth coordinates of the bioprosthetic valve amplitude structure according to the fitting results, such that the overall trend of the surface boundary is smoothened, but the high-frequency changes in fiber bundles and abnormal protrusions are preserved. The OCT amplitude images were then displayed as a 3D (Z-X-Y) structure view and an en-face (X-Y) maximum intensity projection (MIP) of the 3D structure.Results and DiscussionsThe main advantage of OCT is its ability to acquire large field-of-view two-dimensional (2D) tomograms and 3D volume data. From the structural diagram, it can be concluded that the abnormal direction of the fiber bundles on the surface of the valve leaflet fiber layer (see Fig. 3), damaged and folded surface of the smooth layer (see Fig. 4), abnormal defects between the layers (Fig. 4), and cutting defects are valuable information which are suitable for the inspection of valve leaflet defects.ConclusionsThis paper proposes a 3D defect inspection method for bioprosthetic valves based on OCT technology, which can achieve high-resolution, large field of view, and real-time 3D structural imaging. The method is used to perform 3D imaging on the complete bioprosthetic valve stent and valve leaflets and realize the abnormal detection of the fiber layer, smooth layer, interlayer defects, and cutting defects. The obtained results show that the method can realize high-resolution three-dimensional defect inspection of bioprosthetic valves, which is helpful for biological scientists in evaluating valve quality. Further, the method can be used in the field of valve manufacturing and inspection.
ObjectiveRupture of vulnerable plaques with thrombosis is one of the major causes of most acute coronary syndromes. Plaque vulnerability is highly correlated with plaque structure, composition, and mechanical properties, which are typically characterized by thin fibrous caps, lipid-rich necrotic cores, and severe stenosis; in addition, plaque vulnerability is also influenced by the mechanomechanical properties of the vessel wall and plaque. Intravascular photoacoustic (IVPA) imaging is an emerging intravascular imaging modality that can provide submillimeter resolution at penetration depths up to several centimeters and is capable of localizing and imaging lipids with high sensitivity and specificity. Simultaneously, plaque elastography can be performed through the IVPA signals without additional excitation; subsequently, the elastic mechanical properties of plaque can be evaluated. Although intravascular ultrasonography (IVUS) and intravascular optical coherence tomography (IVOCT) have been widely used in the clinical evaluation of plaque diagnosis, the current single intravascular imaging technology cannot assess the vulnerability of atherosclerotic plaques. For a comprehensive diagnosis, the clinicians need to obtain multiple features to fully identify and evaluate plaques. In this study, we proposed and developed an intravascular multimodal system that integrates four imaging modalities, namely, PA, US, OCT, and PAE. The information on plaque based on these four modalities can be obtained from a single probe via 360° rotation and synchronous retraction in one time, which is expected to provide a new interventional imaging method and tools for the understanding, diagnosis, and treatment of atherosclerotic plaques.MethodsThe intravascular PA-US-OCT-PAE imaging system, which integrates the four subsystems of photoacoustic imaging, ultrasonic imaging, OCT, and photoacoustic elasticity, can be used to analyze the macroscopic and microscopic structural information of the blood vessel wall and to specifically identify the lipid components and to perform the elastic mechanics information diagnosis of lipid plaques. The software and hardware designs of each subimaging system and the timing control between the four modalities are shown in Fig. 1 and Fig. 2. The four-modality imaging results are obtained from the only imaging probe by a single-rotation scanning. The structural design of the integrated probe is shown in Fig. 1(b). As shown in the figure, the optical and acoustic paths are placed in parallel, and a miniature ultrasonic transducer with a main frequency of 30 MHz is used for PA signal reception and US signal excitation and reception. A single-mode fiber with an 8° angle end face is used to transmit PA excitation light and OCT detection light and receive the backscattered light of the OCT. A C-lens and coated mirror with a diameter of 0.5 mm are used to focus and deflect the beam so that the focal point of the light is located approximately 2 mm above the ultrasound transducer. The probe house is connected to the torsion coil for torque conduction. The imaging probe assembled with the above design scheme has only a rigid length of 3.6 mm and a diameter of 0.97 mm; this design improves the passability of the probe in tortuous blood vessels. In vitro experiments with simulated samples and in vivo imaging of rabbit abdominal aorta were conducted using the four-modality system with an integrated probe to demonstrate the feasibility of plaque analysis.Results and DiscussionsThe resolution of the PA-US-OCT-PAE imaging system was tested by using eight tungsten wires each with a diameter of 6 μm; the wires were arranged in a spiral form. As shown in Fig. 4, the lateral resolutions of OCT, PA, and US are 20.5, 61.3, and 122.2 μm, respectively. The axial resolutions of OCT, PA, and US are 15.8, 57.4, and 72.5 μm, respectively. The PAE signal, which is attributed to the rising edge of the PA signal, does not contain depth information. Hence, there is no axial resolution, and its lateral resolution is consistent with the PA modality. Further, in vitro experiments on simulated samples of porcine arterial vessels mixed with stents and lipid demonstrate the imaging capabilities of vascular structure identification, lipid detection, and elasticity measurement (Fig. 5). Finally, the in vivo experimental results of atherosclerosis model rabbits show clear vascular PA-US merged three-dimensional (3D) images, OCT 3D images, and PAE results of different sections (Fig. 6). The above results demonstrate the in vivo imaging capability of the intravascular four-modality system, thereby laying the foundation for its clinical translation. However, the system still has some limitations: the PAE signal cannot be extracted from the site where the PA signal is not generated, resulting in the partial absence of the elastic image; in addition, in vivo imaging experiments were only carried on animal models with lipid plaques, and trial data for other types of plaques are lacking.ConclusionsIn this study, a intravascular multimodal PA-US-OCT-PAE imaging system with an integrated imaging probe were developed for the first time. The resolution of the multimodal system was tested by the ultrasonic echo method, and the excellent imaging capabilities of the system were demonstrated. The results of in vitro simulated samples and in vivo rabbit abdominal aorta imaging experiments showed that the PA modality can provide high-contrast images of the lipid distribution in the depth direction. The US modality can reveal the complete vascular structure. The OCT modality can not only evaluate the adherence of the stent but also provide the fine structure of the vessel wall. The PAE modality can provide the elastic mechanics information of the plaque. The information based on the four modalities, which is obtained by only one pullback imaging, is sufficient for comprehensively evaluating the structure, composition, and mechanical properties of the plaques. In conclusion, the intravascular multimodal imaging system is expected to provide a new and comprehensive method for research on plaque vulnerability and help clinicians to effectively diagnose and treat patients with atherosclerotic problems.
ObjectiveThere are a variety of functional proteins localized on the cell membrane that participate in many crucial cellular processes, such as signal transduction and transmembrane transport. The spatiotemporal distribution of specific membrane proteins largely determines their activity states and functions. It is known that the sizes of membrane proteins and the distances between them are both on a nanometer scale. Owing to diffraction limits, traditional optical microscopy cannot provide the spatial distribution of membrane proteins at the single-molecule level. Therefore, imaging techniques with strong specificity and high resolution are urgently required to reveal the precise spatial distribution of membrane proteins. Nowadays, single-molecule localization microscopy (SMLM) offers new opportunities to resolve the detailed distribution information of membrane proteins at the nanoscale, while the great improvement in spatial resolution also presents higher demands for accurate clustering segmentation of images. Density-based spatial clustering of applications with noise clustering (DBSCAN) is one of the most commonly used clustering methods; however, it shows relatively poor performance in clustering segmentation in SMLM images of membrane proteins with heterogeneous density. To address this issue, we propose a novel clustering method using a combination of a multi-step DBSCAN and a hierarchical clustering algorithm. This improved clustering method is based on the traditional DBSCAN method, which combines area threshold analysis and hierarchical clustering.MethodsIn the present work, we improved the traditional DBSCAN method by introducing a variable neighborhood radius and hierarchical clustering to perform precise image clustering segmentation in the original image (Fig. 2). First, we inputted a relatively large parameter (ε1, M1) to perform the DBSCAN calculation. Owing to this relatively large parameter, the excessively discrete points in the original image were removed as noise points. Meanwhile, some of the close-point clusters merged together. Subsequently, the area of each preliminarily identified cluster was calculated and divided by the average area for normalization. Based on the acquired normalized values, we selected an appropriate threshold parameter for extracting clusters with a relatively large area. Subsequently, secondary DBSCAN was performed by the input of a smaller or equal parameter (ε2, M1; ε2≤ε1). For each point cluster extracted in the second step, the calculation was looped from ε2 to ε1. The parameter showing the maximum number of divisible point clusters in the output during the looped process from ε2 to ε1 was selected as the clustering parameter for the next hierarchical clustering. Finally, we combined the above two DBSCAN results to obtain the final clustering segmentation result.Results and DiscussionsWe tested this improved clustering method on both simulated and experimental SMLM data. For the simulation datasets, we chose the D31 and S2 datasets from previous studies as our test objects (Fig. 4). The purity of the improved method on the D31 dataset was 95.64% (86.52% for the traditional DBSCAN method), and the adjusted Rand index was 0.9186 (0.6463 for the traditional DBSCAN method). In addition, the silhouette coefficient and noise ratio were used to analyze the two datasets. Compared with the traditional DBSCAN method, the silhouette coefficient of the improved method significantly increased, and the noise ratio decreased (Table 1). For the S2 dataset, the improved method also exhibited a more accurate segmentation effect than the traditional DBSCAN method. The identification purity of the improved method for the S2 dataset was 95.52% (77.38% for the traditional DBSCAN method), and the adjusted Rand index was 0.9128 (0.6777 for the traditional DBSCAN method). The silhouette coefficient and noise ratio increased and decreased, respectively (Table 1). For the experimental SMLM data, we tested the clustering segmentation effect of the improved method on the uniform, random, and non-uniform SMLM images of membrane proteins (Fig. 5). Similarly, the improved clustering method has a higher accuracy and silhouette coefficient, and a lower noise ratio (Table 1). However, it is regrettable that the time consumption of the improved clustering method is higher than that of the traditional DBSCAN method for both the simulated and experimental datasets (Table 1).ConclusionsBased on the characteristics of the point clusters in SMLM images of membrane proteins, we proposed a novel clustering method that combines area threshold segmentation and multi-step clustering segmentation based on the traditional DBSCAN algorithm. When we applied this method for the image segmentation of simulated datasets as well as experimental SMLM data of membrane proteins, the obtained parameters, including purity, adjusted Rand index, silhouette coefficients, and noise ratio, were generally improved compared with those of the traditional DBSCAN method. On the premise of accurate clustering recognition of super-resolution images and a certain noise reduction ability, the localization information of each cluster can be preserved as much as possible. Our method exhibites a good clustering segmentation ability, especially for SMLM images of membrane proteins with heterogeneous densities. This improved clustering method provides novel insights into the segmentation of membrane protein SMLM images, which is expected to facilitate research into the nanoscale spatial distribution of various membrane proteins.
SignificanceTraditional Chinese physiotherapy, primarily represented by acupuncture and cupping, has been employed for thousands of years to treat musculoskeletal diseases and relieve pain symptoms. Acupuncture is popular worldwide owing to its remarkable curative efficacy and safety. However, the theory of traditional Chinese medicine fails to provide substantial scientific evidence to clarify the mechanisms underlying acupuncture treatment. Moreover, most acupuncture therapies lack effective scientific assessment during treatment, failing to ensure their efficacy and safety. Optical imaging can be used to propagate light in cells and tissues, combine various molecular probes to image organs, and safely obtain robust biological information. Optical imaging is optimal for observing the vascular structures of biological tissues, as well as for monitoring local hemodynamic changes. Optical imaging techniques employed to examine acupuncture primarily include laser Doppler blood perfusion imaging (LDPI), laser speckle imaging (LSI), near-infrared spectroscopy (NIRS), and photoacoustic imaging (PAI). Considerable progress has been made with regard to measuring hemodynamic effects, brain response, therapeutic mechanisms, and the curative effect of acupuncture. However, a systematic summary of these findings is lacking. This review helps readers in the field of traditional Chinese medicine to establish a comprehensive understanding of diverse optical imaging techniques and outline their recent advancements in acupuncture research.ProgressThis review briefly introduces the characteristics of different types of optical imaging and their progress in assessing acupuncture. In addition, their limitations and development directions are summarized.LDPI and LSI use non-contact data acquisition, with advantages such as non-invasiveness and rapid scanning and eliminating certain hidden dangers generated by contact imaging systems. Measuring the improvement in blood circulation at specific sites using LDPI and LSI can assess the efficacy of acupuncture and reveal its therapeutic mechanism. LDPI and LSI can be employed to measure blood flow perfusion of specific regions in real-time, observe changes in internal organs during acupuncture, and verify the correlation between meridians and internal organs. However, the shortcoming of LDPI and LSI should be noted. Owing to the limited penetration depth, research on acupoints and internal organs is only performed on small animals. To determine the microcirculation effect of acupuncture on human viscera, LDPI and LSI should be combined with nuclear medical imaging technologies such as PET.NIRS probes can be easily attached to the skin surface owing to their small size. Currently, NIRS is employed to monitor real-time oxygen levels in the muscle and brain during acupuncture. Owing to its economic advantage, convenience, and safety, functional NIRS (fNIRS) is suitable for examining the changes in hemodynamic parameters in the target area in clinical practice, aiding therapists in effectively evaluating the treatment effects. In recent years, fNIRS has gradually been established as an important supplement to traditional brain functional imaging technologies [such as functional magnetic resonance imaging (fMRI)]. fNIRS has been primarily applied to verify the specificity of acupoints in specific brain regions. However, owing to the skull, human respiration, heartbeat, and other factors, the signal quality of fNIRS has failed to reach the ideal state, accompanied by delayed changes in blood oxygen signals in different regions; this presents a considerable challenge for examining the acupuncture-mediated brain effects using fNIRS. Future developments in fNIRS will focus on improving the filtering algorithm, suppressing physiological interference and random noise, and improving signal delay.PAI combines the advantages of optical and acoustic imaging, easily surpassing the 1 mm penetration depth limit of traditional optical imaging and allowing simultaneous high-resolution and high-contrast imaging. PAI can also monitor blood volume, hemoglobin concentration, blood oxygen saturation, and other tissue indicators while determining structural images. PAI is primarily used to observe the sensitization of acupuncture points, changes in cerebral blood flow perfusion, and cerebral vascular morphology during and after acupuncture. In addition, acupuncture can be used to assist various optical probes, thereby improving the sensitivity and contrast of PAI in the brain. However, traditional PAI requires a couplant to achieve imaging, which is unsuitable for acupuncture. In the future, with the development of air-coupled PAI technology, non-contact PAI will overcome the limitations of traditional PAI and play a role in clinical research assessing acupuncture.Conclusions and prospectsHerein, we summarized the characteristics of different optical imaging methods and their application scope (Table 1). Although optical imaging has facilitated the elucidation of underlying mechanisms and efficacy of traditional Chinese acupuncture therapy, some limitations are known to persist. As the meridian system of the human body is markedly complex, most current studies only select certain formal acupoints. Additional investigations are required to examine changes induced by simultaneously stimulating multiple acupoints and verify the specificity of each acupoint in the human body. In future research, scientists should continue exploiting optical imaging in combination with other imaging methods, such as fMRI or PET, to examine the acupuncture-mediated brain effects, as well as effects on energy metabolism and receptor expression in various regions of the body. This strategy would further reveal the therapeutic mechanism of acupuncture and establish complete guidance, which would benefit a large patient population.
SignificanceThree-dimensional (3D) imaging is an important research direction in microscopy and has been applied to many fields such as biomedicine and engineering science. Typical 3D microscopy techniques, such as laser confocal microscopy and multi-photon microscopy, are based on laser point scanning geometry, and the imaging speed is limited by the scanning speed; therefore, biological samples are likely to be damaged under long scanning durations and high-intensity illumination.Recently, wide-field microscopy with 3D imaging capability has received significant attention. Wide-field microscopy can yield complete two-dimensional imaging simultaneously and affords temporal resolutions higher than spot scanning by two to three orders of magnitude. Additionally, wide-field imaging offers high-quality grayscale images and fewer samples to be damaged, thus rendering it suitable for the real-time observation of living samples. However, conventional wide-field microscopy suffers from defocused backgrounds and low axial resolutions. Owing to the rapid development of computer science and optical technology, various algorithms and techniques for processing wide-field images have been proposed to improve their axial resolutions, thus providing more possibilities for 3D imaging.We focus on three types of rapid 3D wide-field microscopy techniques, i.e., shape of focus (SFF), structural illumination microscopy (SIM), and deep learning-assisted 3D imaging. The SFF technique enables the extraction of focal plane information by processing a series of image stacks and reconstructing the 3D morphology of samples without requiring specific hardware. In SIM, samples are illuminated by phase-shifted light fields with high spatial frequencies, images are captured using a CCD camera, and the in- and out-focus information can be effectively separated using decoding algorithms. Deep learning models can learn the mapping relationship between different types of images from a large amount of data, such as the conversion between wide-field images and confocal images; this is a simple method to obtain high-quality images. The trained model can remove the background information of wide-field microscopic images to improve the axial resolution of imaging, thus facilitating the realization of 3D imaging via wide-field microscopy.It is believed that rapid 3D microscopes based on wide-field imaging will be applied to many fields such as biomedicine, materials science, and precision manufacturing in the near future.ProgressThis paper focuses on three rapid wide-field 3D imaging techniques, namely, SFF, SIM, and deep learning-assisted 3D imaging.In the SFF technique, a focusing evaluation operator is used to calculate and extract the highest focus position of each pixel from a wide-field image stack; subsequently, the 3D depth image of the sample is reconstructed via a recovery algorithm, which is mainly used for surface topography measurement. We investigate the effect of the focused evaluation operator on the calculation results yielded by the SFF technique in different cases. Additionally, we discuss the development and application of the focused topography recovery operator and the optimization of related hardware.Optical sectioning SIM utilizes encoded structured light fields to illuminate the sample and then recovers the 3D information of the sample using decoding algorithms, which can be used for both fluorescent and non-fluorescent imaging. We introduce the theoretical basis of optical sectioning SIM and then propose various rapid decoding algorithms for improving the reconstruction speed. Then, we discuss the development of related techniques and their most recent applications in the field of 3D color imaging.Deep learning-assisted 3D imaging applies the learning ability of neural network models to complete target image tasks, such as the conversion between wide-field and confocal images as well as that between wide-field microscopy and SIM so as to achieve wide-field 3D imaging. We present the theoretical basis of deep learning-related models. Subsequently, we discuss the development and application of deep learning models for conversion between wide-field and confocal images as well as that between wide-field microscopy and SIM, followed by the applications of deep learning for achieving more rapid SIM imaging.Finally, we discuss the current problems and future research directions for rapid 3D wide-field microscopy techniques.Conclusions and ProspectsRapid 3D wide-field microscopy techniques have demonstrated performance improvement either through hardware modification or software assistance. However, these techniques are not perfect. SFF combined with other techniques is expected to benefit deep tissue imaging. The amount of SIM imaging data is two to three times that of the conventional wide-field microscopy, and the imaging speed of SIM can be further improved. Deep learning can be flexibly combined with other technologies. In summary, the potential of wide-field microscopy with 3D imaging capability is yet to be realized. Progress in technology and cross integration will enable the routine use of rapid 3D wide-field microscopy techniques in biomedical laboratories.
SignificanceTwo-photon microscopy (TPM) has been widely used in biological imaging owing to its sub-micron lateral resolution, intrinsic optical sectioning, and deep penetration abilities. TPM enables the observation of cellular and sub-cellular dynamics in deep live tissues within highly complex and heterogeneous environments such as the mammalian brain, thereby providing critical in situ and in vivo information.However, because of the nonuniformity of the refractive index of biological tissues, the laser is distorted and scattered during propagation. Consequently, the focus point becomes a diffuse spot, which leads to a decreased imaging depth and poor resolution of TPM.Adaptive optics (AO) technology was first applied to TPM in 2000, where a genetic algorithm was used to calculate the wavefront distortion and a deformable mirror (DM) was used to correct the aberration introduced by biological samples. Since then, various AO schemes have been developed for a wide range of high-resolution microscopes to advance the development of biological exploration.In this study, the sources and characteristics of aberrations in TPM are examined, and different detection and correction methods in AO are summarized. The different applications of AO in TPM in recent years are comprehensively reviewed.ProgressFor AO technology, wavefront detection methods are generally divided into direct wavefront detection, which uses a wavefront sensor (WS) to detect wavefronts, and indirect wavefront detection, which estimates the aberrated wavefront using iterative algorithms.In 2010, the pupil-segmentation AO method was proposed by Ji et al. This method involves the division of the pupil into several sub apertures and the use of spatial light modulator (SLM) to modulate the wavefront phase, and the imaging resolution of a fixed mouse cortex slice was restored to the near-diffraction-limited (Fig.2). In 2012, Tang et al. proposed an iterative multiphoton adaptive compensation technique that exploits the nonlinearity of multiphoton signals to determine and compensate for distortions and focus light inside deep tissues. The technique was tested using a variety of highly heterogeneous biological samples, and an imaging resolution of approximately 100 nm was obtained (Fig.3). In 2014, Wang et al. adopted a digital micromirror device (DMD) to rapidly modulate the intensity or phase of light rays of multiple pupil segments in parallel to determine the wavefront aberration (Fig.4). Subsequently, Park et al. developed a multi-pupil adaptive optics (MPAO) method in 2017 that allows the simultaneous correction of a wavefront over a field of view of 450 µm×450 µm, thereby expanding the correction area to nine times larger than those of conventional methods (Fig.7). Recently, Rodríguez et al. developed a compact adaptive optics module and incorporated it into both TPM and three-photon microscopy to correct tissue-induced aberrations. They also demonstrated that their technology allows the in vivo high-resolution imaging of both neuronal structures and somatosensory-evoked calcium responses in the spinal cord of mice at great depths (Fig.5).Generally, the indirect wavefront detection system is relatively simple and easy to implement, but it is also time-consuming and has high computational expense. In the typical TPM, the excited fluorescence is limited to a small area near the focus. Such fluorescence points act as a natural guide star for the wavefront sensor to allow the application of direct wavefront detection in TPM. From 2010 to 2013, Cha et al. and Tao et al. used Shark-Hartmann wavefront sensor (SHWS) to detect the emission light and DM to correct the excitation light by injecting foreign fluorescent substances into the sample as a guide star for the in vitro imaging of mice brain tissue (Fig.10). In 2014, Wang et al. proposed descanning technology to accumulate all the transmitted optical signals for wavefront detection and improve the quality and efficiency of direct wavefront detection. The SLM was used to correct the excitation light and in vivo structural imaging was performed on the brain neurons of mice (Fig.11). In 2019, Liu et al. used near-infrared fluorescent dye as a guide star for direct wavefront detection, but replaced the SLM with DM. Structural imaging was carried out on the microvessels and neurons of mice, and an imaging depth of up to 1100 µm was obtained (Fig.12).Compared with indirect wavefront detection, direct wavefront detection is faster and more accurate. However, the optical system for direct wavefront detection is complex, which reduces the imaging signal-to-noise ratio. To effectively use the two detection methods, the real distorted wavefront should be obtained in the optical detection path without a wavefront sensor. In 2017, Papadopoulos et al. proposed the focus scanning holographic aberration probing (F-SHARP) method, which directly measures the point spread function (PSF) of the distorted wavefront using interference technology and corrects the wavefront using the phase conjugation of the PSF (Fig.14). In 2022, Qu et al. combined a conjugate AO with the F-SHARP system and used a phase-locked amplifier to simplify the measurement steps of PSFs, which improves the measurement speed and correction accuracy (Fig.15).Conclusions and ProspectsCurrently, there is an increasing demand for high-resolution structural and functional neuroimaging systems owing to the rapid development of brain science. However, the aberrations caused by tissues are complex, irregular, and rapidly changing and fast aberration detection and accurate correction systems are required. Therefore, it is necessary to use high-performance adaptive elements such as a high sensitivity wavefront sensor and correction elements with a high refresh rate and large compensation range. In addition, fast and accurate compensation algorithms can also improve the AO performance. In summary, the effective combination of various detection, correction and control techniques is the focus of in vivo microscopic imaging, which provides valuable information for scientific research.
SignificanceMultiphoton excitation fluorescence imaging is widely used in the field of biomedical optics and has become one of the most important research tools due to its low invasiveness, strong penetration, high signal-to-noise ratio and high spatial resolution. Photodamage in biological tissues can be caused if excessive photon density or laser power is applied during imaging.While signal-to-noise ratio determines the lower limit of laser power that can be used in multiphoton imaging, photodamage delineates its upper boarders. For in-vivo label-free imaging, due to the small cross section of endogenous fluorophores (≈10-2 GM), the tunability range of laser power between nondestructive imaging and photodamage is very narrow. Therefore, a reasonable laser power is required to ensure that multiphoton imaging with sufficient information can be obtained, and that the cells or biological tissues remain functionally active after long time irradiation. Reducing photodamage and optimizing imaging parameters is one of the major challenges in multiphoton imaging. Photodamage studies are essential to the optimization of imaging parameters.ProgressPhotodamage can be intuitively understood by the concept of ionization penalty in multiphoton bioimaging (Fig. 1). Ionization penalty occurs at irradiance of about 2×1012 W/cm2, where, for a single fluorescence photon emitted from fluorophores, a free electron is produced from water. The chemicals arising from ionization of water molecules can be detrimental to biomolecules and tissues.The underlying mechanisms behind photodamage can be generally divided into photochemical and photothermal effects, as illustrated in Fig. 2. For photochemical effects, they can be further divided into UV-A like photo-oxidation effect and plasma-mediated chemical effect that is wavelength-independent.The severity of photodamage is related to the laser parameters and optical parameters of biotissue. To evaluate the cause of photodamage, the state-of-the-art numerical tools are summarized, including a refined multi-rate-equation model to simulate free electron energy spectrum [Eqs. (4) and (5)], linear and nonlinear heating leading to temperature rise by a single laser pulse [Eqs. (8) and (9)], and heat accumulation by pulse series [Eqs. (11)-(14)].Recent research progresses of photodamage in different tissues and at different wavelengths are analyzed. For two-photon imaging of non-pigmented tissues such as murine intestinal mucosa (Fig. 4), nondestructive imaging can be achieved at average power Pavg≈20 mW, with typical laser parameters of repetition rate fPRF≈80 MHz, wavelength λ≈800 nm and pulse duration τL≈100 fs. Photodamage occurs when ≥2 times imaging power at focus is used. In contrast, for pigmented tissue such as murine retina (Fig. 7), photodamage occurs at average power as low as 3.5 mW with fPRF≈80 MHz. Simulation results show that photodamage in non-pigmented tissue is mediated with laser-induced low density plasmas (Fig. 6), whereas photodamage in pigmented retina is mainly driven by heating (Fig. 8).For three-photon imaging of deep murine brain tissue (≈1 mm) using typical laser parameters λ≈1.3 μm, fPRF≈1 MHz, a non-zero chance of photodamage is observed for laser power Pavg≥150 mW (Fig. 9). Immunostaining as well as Monte-Carlo simulation results indicate that linear absorption and heating are likely to cause the photodamage. However, multi-rate-equation modeling shows that laser-induced plasma-mediated effects may be involved as well (Fig. 10).Conclusions and ProspectsIn this review, we analyze photodamage in pigmented and non-pigmented tissues in multiphoton imaging. We come to the conclusion that photochemical effects are dominant in two-photon imaging of pigment-free tissues, while photothermal effects play a leading role in two-photon imaging of pigmented tissues. For three-photon imaging of deep murine brain tissue, photodamage is likely to arise from synergistic photochemical and photothermal effects.Fully using the photon budget without photodamage is still a big challenge in multiphoton imaging. Traditional optimization model is time-demanding. Recently, with the booming development of machine learning, it has been applied to the optimization research of super-resolution optical microscopy. Subsequent combination of photodamage threshold database and machine learning can be a new direction to achieve online, automatic optimization of imaging parameters for multiphoton imaging.
SignificanceThe brain is one of the most complex systems, the culmination of evolution over billions of years of life. But until now we have not been able to accurately describe the mechanism of memory, thought and consciousness. Due to the lack of understanding of the structure and function of the brain, we have no effective drugs and treatments for neurological diseases such as schizophrenia, epilepsy, Alzheimer’s disease, and Parkinson’s disease. The structure of the brain is extremely complex and can be divided into different levels, such as brain lobes, neural circuits, neurons, synapses, and even molecules. The brain’s powerful function stems from its huge number of nerve cells and their complex interconnections.The mapping of whole-brain mesoscopic neural connections in model animals such as mice requires technical tools that can achieve large-scale acquisition of high-resolution three-dimensional data in the centimeter scale. Optical imaging methods can achieve sub-micron resolution in lateral direction and can realize “optical sectioning” by various means, which have the natural advantage of observing neural circuits at the mesoscopic level. In this review, we summarize the various kinds of whole-brain optical imaging methods developed in recent years and look forward to future technological development.ProgressDue to the scattering and absorption of biological tissues, the imaging depth of traditional optical methods is limited, and only tens of microns to hundreds of microns of the shallow layer of mouse brain can be imaged. To break through the limitation of imaging depth in biological tissues and achieve high voxel resolution and large-scale 3D imaging, optical microscopy must be combined with histological methods (Fig. 1).Tissue clearing based whole-brain optical imaging methods are technologies that first clear biological tissues to improve optical imaging depth, and then use light-sheet fluorescence microscopy (LSFM) for rapid imaging (Fig. 2). For a whole mouse brain sample, rapid imaging of micrometer resolution can be completed in a few hours or less by LSFM. LSFM can achieve good optical sectioning with low photobleaching and phototoxicity. However, the imaging resolution of LSFM decreases significantly with the increase in the depth of the sample, and the resolution is lower when a large sample is fully imaged.Mechanical sectioning based whole-brain optical imaging methods are a combination of optical sectioning and tissue cutting. After acquiring the images of the shallow part of sample each time, the sample surface is cut off with a knife. Through the continuous cycle of “imaging-cutting” process, the 3D fine structures of centimeter-size sample can be obtained. Representatives of this technology include serial two-photon tomography (STP), block-face serial microscopy tomography (FAST), and micro-optical sectioning tomography (MOST) series technologies.STP combines high-speed two-photon imaging with vibrating sectioning, and can realize Z-interval sampling imaging of mouse brain (Fig. 3). With the use of resonant scanning galvanometer and higher excitation light intensity, the speed of STP imaging is further improved. By optimizing the mouse brain clearing method, the imaging depth can be improved to more than 200 μm. Finally, the high-resolution mouse brain data of 0.3 μm×0.3 μm×1 μm can be obtained within 8-10 days.FAST uses spinning-disk confocal technology to image shallow tissues within a thickness of about 100 μm, and can complete monochromatic imaging of whole mouse brain within 2.4 h at a voxel size of 0.7 μm×0.7 μm×5 μm (Fig. 4). Sparse imaging and reconstruction tomography (SMART) uses low-resolution imaging results to determine whether there is fluorescence signal in the current region to narrow the next scan region, enabling rapid fine imaging of transparent sparse labeled mouse brain.MOST uses a diamond knife to perform continuous sectioning of 1 μm thickness of resin-embedded mouse brain samples, and at the same time performs line scanning imaging of sample sections on the blade edge (Fig. 5). The mouse brain Golgi staining dataset with a voxel size of less than 1 μm3 has been acquired for the first time. Structured illumination fluorescence micro-optical sectioning tomography (SI-fMOST) adopts structured illumination method to achieve high-throughput imaging of the sample block-face by mosaic scan stitching (Fig. 5). Combined with real-time staining, dual-color imaging of mouse brain can be completed in 3 days with voxel size of 0.32 μm×0.32 μm×2 μm. High-definition fluorescence micro-optical sectioning tomography (HD-fMOST) uses the Gaussian intensity distribution of line illumination as a natural modulation, and can effectively remove background through simple subtraction (Fig. 5). The MOST series technologies combine embedding brain samples, micro-optical imaging and automatic precision cutting to form a unique technical system. The collected datasets are characterized by excellent integrity, high resolution, and good quality.Conclusions and ProspectsThe whole-brain imaging technologies achieve unprecedented resolution, imaging speed and imaging range, making it possible to study the whole-brain circuit network. These technologies have gradually become a powerful tool in neuroscience and will continue to develop into more universal research tools, thus showing more application value.To map neural circuits, it is necessary not only to develop imaging technology, but also to develop methods such as sample labeling and preparation, massive data storage, processing, and visualization. In recent years, the development of various virus tracing tools has promoted the labeling of neural circuits. But how to process and analyze massive whole-brain imaging data and extract knowledge from it is likely to become a bottleneck.As the closest species to humans, non-human primates are of great value for the study of cognitive behavior, disease mechanism and treatment. However, the weight of macaque brain is about 200 times that of a mouse brain, which poses great challenges to sample labeling preparation, imaging technology and big data processing.Through interdisciplinary cooperation, whole-brain optical imaging will further flourish, demonstrate its unique application value in neuroscience, promote our knowledge and understanding of the brain, and contribute to the development of artificial intelligence technology.
ObjectiveOptical coherence tomography (OCT) is a widely used imaging technique in retina research, with the spectrometer being a crucial component that determines the performance of spectral domain OCT (SD-OCT). While there are commercial spectrometers and systems available with a variety of options, they are often expensive and not customizable for specific light sources and applications. Thus, independently developing spectrometers and OCT systems could provide a better alternative. The calibration of a spectrometer is typically complex because it requires a standard light source, such as a mercury lamp, that must meet the specific requirements for calibration, including accurate spectral characteristics. Additionally, use of such a light source demands certain technical and operational expertise. Therefore, this paper proposes a practical calibration method for an OCT spectrometer based on a common OCT algorithm. As a result, the need for a standard light source is eliminated, and hence OCT spectrometer calibration is simpler and easier.MethodsIn this study, an SD-OCT system was built, incorporating a supercontinuum laser as the laser source. The corresponding wavelength range is 800?950 nm using filters. The low coherent light emitted by the laser is split into two beams through a fiber coupler. Each beam enters the sample arm and the reference arm, respectively. In the sample arm, the light passes through a two-dimensional galvanometer, generating a scanning beam on the mouse retina in this work. The power of the beam at the mouse pupil was approximately 600 μW, with the beam diameter of 0.93 mm. To minimize chromatic aberrations, the lenses used in both the sample arm and reference arm were paired appropriately. The reflected beams from the sample arm and the reference arm combine and interfere on a custom-built spectrometer. The spectrometer includes a transmission grating, a line CCD camera, and other optical devices. For spectrometer calibration, a mirror is placed at the retina plane, reflecting light back to the spectrometer similar to the reference arm setup. To achieve accurate calibration and performance analysis, the optical power from both arms was adjusted using irises to achieve similar intensity. The interference fringes at different imaging depths are then captured by the camera, facilitating subsequent calibration procedures. The calibration process involves synchronously optimizing the peak value and full width at half maximum of specular reflections collected at these depth positions via manual tuning of difference parameters. Finally, OCT imaging experiments on ten mice were conducted to validate the performance of the spectrometer.Results and DiscussionsThe quantitative analysis results of the spectrometer are presented in Fig. 5. Figure 5(a) shows the spectral curve of the light source directly measured by the spectrometer. In Fig. 5(b), the k-value linearization curve is displayed. The spectral data obtained after Fourier transform is shown in Fig. 5(c), with the peak range of 127 dB?104 dB. To determine the corresponding relationship between CCD camera pixels and spatial distance in A-scan, the position of the peak in Fig.5(c) was extracted and correlated with the actual moving distance of the displacement platform. The calculated relationship between pixels and actual spatial distance was determined as 2.65 μm/pixel in the air, as depicted in Fig.5(d). The spectral data in Fig.5(c) was further Gaussian fitted and multiplied by the above obtained relationship to determine the maximum and minimum axial resolutions in the air of the system, which are 4.14 and 2.72 μm, respectively. The axial resolution change curve remains relatively stable within the imaging range, as demonstrated in Fig.5(e). Additionally, the sensitivity change curve [Fig.5(f)] was realized by connecting the data peaks in Fig.5(c) with a polygonal line. To evaluate the practical application of the spectrometer in mouse retina imaging, 1000 B-scan images were collected at the same position using the OCT system. Each B-scan comprised 1083 A-scans, with the A-scan rate of 100 kHz. The acquired image data were then aligned, averaged, and contrast-enhanced using ImageJ. The mouse retina OCT images are presented in Figs.6(b) and 6(c). To analyze the retina’s structure, the profile of each retinal layer was obtained by averaging the image in the horizontal direction, as depicted in Fig.6(d). Based on this profile, the thickness of each layer of tissue was measured. The comparison results for the thickness of each layer of the mouse retina are detailed in Table 2, demonstrating the successful implementation and performance of the spectrometer in mouse retina imaging. As a result, valuable insights are provided regarding the retinal structure, with potential application in further research investigations.ConclusionsTo address the demand for high-resolution imaging of the mouse retina in basic science research, a specific SD-OCT system was designed and constructed. The system is based on a customized broadband spectrometer. Herein, the design process of the spectrometer is introduced comprehensively, and an alternate optimization approach to its calibration is proposed based on a few key performance metrics. A notable advantage of this calibration approach is that accurate calibration of a spectrometer is achieved without relying on a standard light source. This streamlined process significantly simplifies the calibration procedure, making it more efficient and cost-effective. Overall, the method offers a practical and convenient solution for optimizing OCT systems. In conclusion, the SD-OCT system presented in this paper, with the custom broadband spectrometer and novel calibration approach, is a practical and convenient tool for achieving high-resolution imaging of the mouse retina in basic science research.
ObjectivePhotoacoustic computed tomography (PACT) is an important photoacoustic imaging modality. Compared with photoacoustic microscopy, PACT can detect biological tissues located several centimeters deep without external contrast agents. Equipped with a multi-channel data acquisition card, PACT has the potential for high-speed imaging under a large field of view and is currently used in clinical and preclinical applications, such as whole-body imaging of small animals and human organs. However, skin tissue contains a lot of melanin, and the high-intensity photoacoustic signal from the skin covers the deep subcutaneous tissue information during the imaging process, hindering the en-face display and analysis of the photoacoustic image of the region of interest. Existing works have successfully removed most of the skin signals in photoacoustic images, but there are still some existing problems: (1) most of them are based on photoacoustic microscopic images of shallow tissues or directly extracted vascular structures in the background; the skin removal of deep tissue PACT images has not been reported; (2) the current pixel-level manual labeling takes a lot of time, and there are shortcomings of low extraction accuracy and low efficiency; (3) owing to reconstruction artifacts and changes in light intensity, the signal amplitudes of the skin area are uneven, and there exists many small segments that cannot be distinguished from the background, which increases the difficulty of extracting a complete and continuous skin signal.MethodsConsidering the continuity of the skin tissue and the uniformity of the thickness of the local imaging area, this study proposes a U-shaped deep learning (DL) model that combines multi-scale perception and a residual structure (MD-ResUnet) to automatically remove skin areas in PACT deep tissue photoacoustic images. The introduction of the residual structure in this model can integrate low- and high-level feature information to prevent model degradation, and the multi-scale dilated convolution blocks can increase the continuity and integrity of skin extraction. In the skin segmentation task, a single-type skin region label was proposed as the ground truth, which significantly reduces the complexity of data annotation, compared with the previous pixel-level multi-type annotation. Subsequently, an algorithm of skin integrity fitting and skin mask generation was designed based on the extracted binary image of the skin, to realize the automatic removal of the skin signal in the PACT image. A total of four PACT datasets were used in our experiments, two of which were used for model optimization and two for experimental verification.Results and DiscussionsThe photoacoustic images of the peripheral blood vessels of human legs from PACT verified the correctness and effectiveness of the proposed method on high-precision extraction and removal of skin tissue. In the task of skin segmentation, the comparative experiments with the existing network models of Unet and Res-Unet, show that the DL model MD-ResUnet proposed in this study can fit most of the narrow skin segmentation gaps, effectively shorten the large segmentation gaps, and the extracted skin is overall more accurate, smooth and continuous (Fig. 4). Compared with the existing skin removal works, the deep learning method proposed in this study can thoroughly remove the skin signal and restore a more realistic and clear deep tissue structure (Fig. 5). Quantitative analysis shows that the reconstruction error of the skin-free image has dropped by 50%?70%, and the peak signal-to-noise ratio is averagely increased by 4.5 dB (Table 2), which may provide an effective method for the high-definition display of deep tissue PACT images.ConclusionsThis study proposes a novel skin removal method for PACT deep tissue images with skin region segmentation as the core and designs a new U-shaped DL network MD-ResUnet to achieve the skin segmentation task. The proposed single-class skin-area labeling method significantly reduces the complexity of data processing, and the boundary fitting and mask generation methods realize the complete removal of skin areas, providing an effective method for high-quality deep tissue image generation in PACT. However, the network model proposed in this study cannot yet achieve fully continuous skin-region extraction, and there are still partially disconnected skin gaps. In addition, the experiment in this study is based on the imaging of the peripheral blood vessels of the human leg. The surface of this tissue is relatively regular, and the overall shape of the skin is arc-like, which is convenient for DL to grasp its overall structure features. For imaging tissues with complex surfaces, such as fingers and wrists, the surface shape of the skin is variable, and there will be more significant illumination differences in the same image frame, resulting in increased uneven skin area signals. In the future, we will explore advanced DL network models to implement the extraction of fully continuous skin surfaces in PACT images.
ObjectivePolarized diffraction images (p-DIs) can provide a wealth of information about the morphologies of scatterers, making them a valuable tool for use in a variety of applications, including the characterization of biological cells and tissues. However, most studies on biological cells have chiefly relied on qualitative analysis, which is achieved through the analysis of patterns of p-DIs for cell clustering. Although qualitative analysis can provide major insights into the morphologies and characteristics of cells, it may not always provide accurate quantitative data about the sizes and shapes of cells, which is critical for some applications. Although quantitative studies on the refractive indices and sizes of cells have been conducted, these investigations have typically been based on the assumption that cells are perfectly spherical. Accordingly, this assumption may not hold true for many types of cells that exhibit non-spherical shapes. Based on a texture analysis of p-Dis, this study conducted quantitative analyses on the characteristic parameters of yeast monomers and budding yeast, where the results are shown to be consistent with those obtained using traditional microscopy methods. This approach can provide insights into the quantitative analysis of non-spherical cells based on light-scattering techniques.MethodsA systematic study on p-DIs with scattering angles of 60°?120° and azimuth angles of 150°?210° of 1197 yeast monomers and budding yeast was conducted using optical models established based on the discrete dipole approximation theory (DDA). Excluding the assumption that the two short aixs of yeast cells are equal, all parameters of these optical models were obtained through microscopy. The experimental p-DIs of 25000 Ale and Lager yeasts were obtained using polarization diffraction imaging flow cytometry. The Fourier spectrum and gray-level co-occurrence matrix (GLCM) parameters of all p-Dis, including those derived from a simulation and experiment, and the depolarization coefficient of p-DIs of yeast monomer were calculated. A regression model was used to establish the quantitative relationship between image feature and cellular structural parameters, such as the sizes of the short aixs of yeasts, the aspect ratio of yeast monomer under different short axis sizes, and short axis ratio of bud yeast to mother yeast. In addition, a statistical correlation between characteristic parameters of yeast cells and GLCM parameters was investigated, and a support vector machine (SVM) classifier was trained based on simulated p-DIs to classify the yeast monomers and budding yeasts in the GLCM parameter space.Results and DiscussionsThis study finds a significant statistical correlation between GLCM parameters and short axis ratio of bud yeast to mother yeast (Fig.6). Therefore, for a statistical analysis of budding rates, the SVM model shows an accuracy of as high as 98.1% [Fig.7(a)]. Moreover, the budding rates of yeasts as calculated by the SVM classifier and microscopic count method are found to be highly consistent [Fig.7(b)]. Further analysis shows that the adjusted R2 as determined by the multiple regression equation is 0.86, indicating that the regression model has very high statistical significance and good predictability. In addition, a power law relationship derived from the nonlinear least squares fit between the normalized spatial frequency along the θ direction and short axis of yeasts is obtained with an R2 value of 0.9986 and narrow 95% prediction interval, indicating that the precision and reliability are satisfactory [Fig.9(a)]. With the aid of this power law relationship, the statistics of the short axis size distribution of yeast monomer based on experimental p-DIs are realized with an error of 7.4% [Fig.9(b)]. The correlations between the aspect ratio of yeast monomer under different short axis sizes and the depolarization coefficients were also analyzed. We find that when the short axis sizes vary in the range of 5?8 μm, the changing trends of the depolarization coefficient with the aspect ratio of yeast monomer under different short axis sizes are consistent and assumed to be a Gaussian function (Fig.10). These results indicate that polarized diffraction imaging technology has promise in terms of quantitative analysis of the structural parameters of non-spherical cell models.ConclusionsA comprehensive numerical and experimental study on the polarized diffraction characteristics of yeast cells is conducted. We develop a yeast cell structural parameter prediction model based on texture features extracted from p-DIs. This prediction model can accurately and rapidly predict yeast cell structural parameters such as short axis size, aspect ratio, and bud size based on given p-DIs. The accuracy and reliability of the model were validated through comparison with actual measurement data. The ability to predict yeast cell structural parameters in a fast and accurate manner is of great significance for the study of cellular morphology and may have major implications for the development of new diagnostic and therapeutic tools.
ObjectiveAs a nonlinear optical imaging technique that offers high spatial resolution and high penetration depth, second harmonic imaging holds great promise for clinical diagnosis and various applications in the biomedical field, because it overcomes photobleaching and saturation absorption owing to energy absorption, which are commonly encountered in fluorescence imaging. Second-harmonic generation (SHG) is a nonlinear optical process in which two identical photons interact with a nonlinear material and are effectively converted into a single photon with precisely twice the frequency of the incident beam. In biologically relevant SHG imaging, the predominant approach has traditionally relied on the use of exogenous dye markers or endogenous proteins with a relatively low SHG efficiency. In fact, numerous studies have demonstrated strong and photostable SHG signals generated by inorganic crystalline materials. However, most of these inorganic crystalline materials contain heavy metals and have relatively large sizes (~100 nm in diameter). Recently, silicon quantum dots (SiQDs) were developed and have attracted growing interest owing to their remarkable properties, such as aqueous solubility, low cytotoxicity, high quantum yield, and exceptional stability against photobleaching. However, only few studies have investigated the generation of SHG signals from SiQDs without structural reconstitution, which has great potential for advanced optical applications, particularly in the field of SHG imaging. In this study, we developed polyethylene glycol (PEG)-coated SiQDs, an asymmetric material with a high nonlinear optical effect, as second-harmonic probes. To enhance the biological affinity and reduce the surface oxidation of the SiQDs, we modified their surface with PEG and investigated the imaging effect of PEG-coated SiQDs as a biological probe for second-harmonic wave imaging in HepG2 cells. Compared to two-photon fluorescence imaging, the second-harmonic imaging technique based on PEG-coated SiQDs provides more reliable and stable results. This finding can promote the future applications of SiQDs in molecular imaging, drug delivery, and stem cell therapy. By combining the advantages of the SHG dye, which has good biocompatibility and extremely low cytotoxicity, and the SHG inorganic crystalline materials with the photostability of the crystal structure, our SiQDs are expected to become the primary choice among many probes. We labeled hepatocellular carcinoma (HepG2) cells with non-functionalized SiQDs for cell imaging using SHG.MethodsFirst, the nonlinear material used in this study, PEG-coated SiQDs, was synthesized by directly reducing the precursor with silicon-oxygen bonds and then modifying with organic ligands. The morphology and chemical composition of the SiQDs were characterized through transmission electron microscopy (TEM) and energy-dispersive X-ray spectroscopy (EDS). Furthermore, the physical mechanism behind the strong SHG of SiQDs was examined using finite-difference time-domain (FDTD) numerical simulations. The second harmonic characteristics of the SiQDs were then evaluated experimentally using a custom-built setup. Finally, to verify the feasibility of using the PEG-coated SiQDs in cell labeling and imaging, SHG imaging studies on HepG2 cells were conducted using confocal microscopy.Results and DiscussionsThe TEM image of the PEG-coated SiQDs [Fig.1(a)] reveals that they are approximately spherical and have an average diameter of (2±0.5) nm. The excellent second-order nonlinear effects of these SiQDs were verified both theoretically and experimentally. By scanning the SHG signals of the SiQDs [see Fig.4(b)], we confirmed that they exhibit strong and stable SHG signals. Furthermore, we used these SiQDs to perform nonlinear optical imaging of HepG2 cells. Confocal microscopic visualization of the HepG2 cells treated with PEG-coated SiQDs confirmed the excellent tracking and imaging ability of the SiQDs (Fig.5). Furthermore, a TPL scan of the cells incubated with SiQDs demonstrated the advantages of SHG imaging over TPL imaging (Fig.6). Overall, the PEG-coated SiQDs serve as stable and reliable biological probes, significantly improving the image contrast compared with that of two-photon fluorescence imaging. The advantages of SHG imaging, including the absence of photobleaching, blinking, and saturation absorption, are highlighted. In addition, the intensity of the SHG signal produced by the PEG-coated SiQDs is 100 times higher than that obtained in two-photon fluorescence imaging [Fig.6(m)]. These results indicate that SHG imaging based on PEG-coated SiQDs has great potential for a wide range of applications in biomedical imaging and other related fields.ConclusionsThis paper presents a method for preparing PEG-coated SiQDs and their application in cell imaging. The PEG-coated SiQDs have a dynamic fluid diameter of only (2±0.5) nm, and the PEG molecules on their surface enhance their biocompatibility and show no apparent toxicity. The main innovation lies in the exceptionally strong and stable SHG signals exhibited by SiQDs. The SiQDs were employed as biological probes for SHG imaging of human liver cancer cells (HepG2). The advantages of SHG imaging, including the absence of photobleaching, blinking, and saturation absorption, were highlighted by comparing the results with those of two-photon fluorescence imaging. Thus, SiQDs can serve as highly biocompatible photosensitizers without causing toxic side effects and thus have promising prospects in biomedical applications.
ObjectiveIn all blind eye diseases, fundus disease remains the primary cause of irreversible visual loss, significantly impacting visual acuity. In severe scenarios, this results in a higher prevalence of blind fundus disease. Many fundus diseases manifest in the eye periphery. If the lesion does not affect the macular area, patients often overlook early lesions since their visual acuity remains largely unchanged, presenting no symptoms. During examinations, standard fundus-imaging equipment fails to visualize the peripheral area of the fundus due to its limited imaging field of view. Once the lesion impacts the central macular area of the fundus, treatments become challenging, and outcomes are generally unfavorable. As such, early examinations play a crucial role in preventing and treating fundus diseases, underscoring the need to innovate instruments that image the retina, encompassing the fundus periphery. Traditional fundus photography has a field of view limited to 30°?50°. Even utilizing multi-region fundus image stitching only marginally expands the fundus imaging area, concentrating the imaging on the posterior pole. Conversely, laser scanning confocal fundus imaging offers superior clarity and contrast, enabling non-mydriatic fundus imaging even in patients with constricted pupils and facilitating real-time dynamic observations of fundus image changes. Ultrawide-angle fundus imaging rooted in laser scanning confocal imaging emerges as a significant advancement in fundus imaging. We anticipate that our alignment method and study findings will inform the design of cutting-edge ophthalmic examination devices.MethodsTo thoroughly image the peripheral area of the fundus, we explore the fundus line-scanning imaging technology and construct an ultra-wide-field confocal laser line-scanning fundus imaging system. Initially, we devise a comprehensive optical pathway for the system. For achieving ultra-wide-angle, high-resolution dual-mode imaging, it is essential to design the geometrical optical parameters of the components within the respective mode, ensuring that the parameters satisfy the dual-mode imaging requirements simultaneously. We commence by establishing the overarching framework of the optical system, which incorporates the parameter design for ultra-wide-field, high-resolution dual-mode imaging. This is followed by Zemax simulations and image quality optimization for the system detection and imaging sections. Components are chosen based on these parameters, leading to the construction of the experimental system. By utilizing the pixel boundaries of the target surface in the area camera, we are able to achieve line scanning dual-mode confocal imaging of fundus through the creation of virtual slits. Once the theoretical design phase concludes, we employ Zemax software to simulate the system detection optical path, optimize this path, and validate the system design metrics and viability. The camera pixel boundary forms a virtual confocal slit, facilitating line scanning dual-mode confocal imaging of the fundus. We then assess the actual field of view of the system, resolution, and imaging capabilities.Results and DiscussionsThe designed laser line-scanning ultrawide-angle confocal fundus imaging system in this study realizes ultrawide-angle, high-resolution dual-mode imaging by simply switching the eyepiece lens (Fig. 1). After parameter design and simulation (Table 1), the commercially available lenses for scanning, lighting, and imaging objectives fully meet the system requirements, reducing the system design cost. In the ultrawide-angle mode, the system actual field of view reaches 136.3°, achieving ultrawide-angle imaging (Table 3). In the high-resolution mode, the system equivalent conversion fundus resolution stands at 8.5 μm, accomplishing high-resolution imaging (Fig. 9). We conduct ultrawide-angle mode imaging, ordinary fundus camera photography, and high-resolution mode imaging on the simulated eye, and the system dual-mode imaging effect proves significant (Fig. 10).ConclusionsThis study offers a method for achieving ultrawide-angle confocal imaging of the fundus based on line scanning. The system employs a Powell prism in conjunction with a cylindrical lens to produce an ultra-long and ultra-fine laser line beam. It utilizes the pixel boundary of the camera target surface to establish virtual slits, achieving confocal fundus imaging. This effectively diminishes the interference of non-focal plane stray light on the fundus image. The system possesses both an ultra-wide-angle fundus imaging mode and a high-resolution imaging mode. Theoretical calculations and experiments indicate that the field of view in the ultrawide-angle mode is 136.3°, and the actual resolution in the high-resolution mode is 8.5 μm. Grounded on the experimental results, the proposed laser line-scanning fundus imaging method proves feasible. It effectively achieves ultrawide-angle fundus imaging and serves as a reference for the development of related instruments.
ObjectiveFunctional near-infrared spectroscopy (fNIRS) is currently widely applied in clinical research on functional brain activity states because of the advantages of fNIRS over conventional in vivo brain function detection techniques. fNIRS is a non-invasive and non-radiative technique that is resistant to electromagnetic interferences, provides a reasonable temporal/spatial resolution, and facilitates direct detection of blood oxygen metabolism. As an emerging reconstruction strategy for fNIRS, diffuse optical tomography (DOT) can complete the 3D reconstruction of optical parameters based on accurate photon transport models and can significantly improve the quantitative accuracy and spatial resolution of typical optical tomography techniques. Owing to the reflection measurement geometry of DOT, the detection data are affected by superficial physiological interferences (cardiac pulsation, respiration, and low-frequency oscillations) and random noises (photon-shot and instrumental noises) that originate from the scalp-skull layer; these interferences and noises affect the accuracy and precision of the reconstruction results. In addition, owing to limited boundary measurements, the inverse problem of the DOT has a non-negligible ill-posedness. Thus, handling the ill-posedness of the DOT inverse problem and suppressing physiological interferences and random noise are critical tasks in fNIRS-DOT neuroimaging. In this study, a model-based reconstruction-informed and deep learning approach, composed of a semi-three-dimensional (S3D) DOT and deep convolutional encoder–decoder neural network (DCNN), is developed to improve the reconstruction accuracy and suppress physiological interferences and random noises.MethodsFirst, an S3D-DOT model is developed based on the properties of near-infrared light activation information distribution in the depth direction and reasonable assumptions about the structural characteristics of the brain. The S3D-DOT model can help in reducing the number of unreconstructed parameters, handle the ill-posedness of the DOT inverse problem, and preliminarily discriminate perturbation maps corresponding to the surface and cerebral-cortex (CC) layer. The preliminary reconstructed image is then used as an input to the subsequent DCNN model, which is composed of two parts, viz. a decoder network and an encoder network. The DCNN model can collect the spatial feature information of the image, effectively separate the activation and interference information, and accurately reconstruct the activation feature in the CC layer map. In general, the proposed model-informed deep-learning architecture is supported by physical models, exploits the spatial-information-extraction capability of convolution and encoding-decoding networks, and can provide highly quantitative and accurate reconstruction results in different application scenarios.Results and DiscussionsThe structural design of the network, parameter selection process, and training process are described in detail. To verify the effectiveness of the proposed method, numerical simulations and phantom experiments are conducted using the fNIRS-DOT system. The final reconstructed images of the proposed method are compared with those obtained using the algebraic reconstruction technique (ART), and appropriate quantitative evaluation indices are selected for the computational analysis. The results of the numerical simulation experiments at specific time points show that the DCNN can effectively suppress the effects of physiological interference and random noise and improve the reconstruction accuracy, with a mean structure similarity index (SSIM) value of >0.998 (Fig.3). DCNN is more advantageous than the ART at a weak excitation time point, and the corresponding time required for reconstruction is significantly less. Subsequently, the performance of the DCNN model is examined under strong noise interferences. The corresponding results demonstrate that the conventional method cannot accurately reconstruct the excitation distribution under these conditions, whereas the proposed algorithm can still guarantee the validity of the reconstruction results (Fig.4). Additionally, the reconstruction capability of the DCNN in complex scenarios is verified through dual-target simulations (Fig.5). Furthermore, practical applicability of the proposed method is preliminarily examined through phantom experiments. The results indicate that the method can accurately filter random noise; however, the reconstructed image is still affected by physiological interferences when its relative intensity is large (Fig.7). Finally, a 3D deep convolutional encoder-decoder neural network (3D-DCNN) model is proposed to enhance the network’s ability to utilize temporal–spatial information and reasonably predict the changes in the excitatory brain regions. The results of the numerical simulation experiments prove that the 3D-DCNN model is more sensitive to small absorption changes and can accurately reconstruct the complete time courses of the average absorption perturbation in the activated region (Fig.9).ConclusionsIn this study, a model-based reconstruction-informed and deep learning approach is developed for enhancing the fNIRS-DOT performance. This proposed approach adopts the S3D-DOT model and DCNN to reduce image artifacts induced by physiological interferences and random noise. This method requires less hardware devices and provides an explicit physical explanation, an excellent accuracy and generalization for different scenes, and a fast reconstruction speed. To assess the effectiveness of the proposed method, a series of preliminary numerical simulations and phantom experiments are conducted, and the results are compared with those of the traditional reconstruction method. The results show that this method can significantly improve the quantification of images, greatly reduce the reconstruction time, and facilitate an excellent generalization, thereby providing an important new reference for dynamic fNIRS-DOT imaging.
ObjectiveCurrently, various super-resolution imaging technologies can surpass the Abbe diffraction limit, thereby improving imaging resolution to several tens of nanometers. This provides biologists with an effective tool for investigating biological structures and their functions on a novel scale. Among these, single-molecule localization techniques such as photoactivated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM) yield the highest resolution. Traditional fitting-based methods, such as single-emitter localization (SE) and multi-emitter localization (ME) algorithms, employ fixed-size sliding windows to select the fitting areas. However, this was found to lead to an inadequate use of the prior emitter recognition information during the emitter localization stage in this study, thereby resulting in diverse advantageous density ranges and different artifact forms of SE and SM. The SE results are distorted by truncates near the emitters, which are generated by the fixed sizes of the fitting areas, whereas the ME suffers from an inappropriate fitting number. In summary, a self-adaptive mixed-emitter single-molecule localization algorithm (SM) that can adaptively determine the fitting area and fitting number is proposed in this study. Consequently, compared with the SE and ME algorithms, the images reconstructed by the SM algorithm exhibit a superior resolution and contrast over the complete density range on both simulated and experimental data.MethodsThe complete SM algorithm comprises several steps. First, an SNR binary map that can shrink and expand with the power of noise was generated based on the original image. Subsequently, the SNR binary map was combined with the local maxima for emitter recognition, and the sliding window and fitting number were generated using the SNR binary map. The center and size of the generated sliding window were then determined based on the center position and size of the connected domain, respectively, whereas the fitting number was obtained from previous emitter recognition results. Subsequently, maximum likelihood estimation (MLE) or least squares (LS) fitting was performed in each fitting area to obtain the subpixel positions. Finally, the performance of the SM algorithm was investigated using simulated and experimental data.Results and DiscussionsUnder a low or high labeling density, the SM algorithm can effectively reduce crosstalk and mismatch errors, which promotes the recovery of super-resolution images closer to the synthesized benchmark images compared to those recovered by the SE and ME algorithms (Fig. 1). For a low labeling density, the SM algorithm exhibits a slightly better precision, recall, Jaccard index, and RMSE than the SE algorithm, and significantly superior results compared to those of the ME algorithm. With an increasing labeling density, the SM algorithm is marginally inferior to the ME algorithm in terms of the precision, recall, and Jaccard index, but is still significantly better than those of the SE algorithm. In terms of the RMSE, the SM and SE algorithms exhibit comparable localization errors, which are both worse than those of the ME algorithm [Figs. 3(a)?(c)]. Quantitative comparisons between the synthesized benchmark images and super-resolution images recovered by the different algorithms are performed using three indicators: PSNR, SSIM, and RMSE. The SM algorithm produces images with a higher similarity to the ground truth, as indicated by all three indicators (Table 1). In addition, it also successfully restores the structure with an interval of 20 nm, which is not achieved using the SE and ME algorithms [Figs. 3(d)?(e)]. On the α-tubulin dataset labeled as Alexa Fluor 647, the SM algorithm outperforms both the SE and ME algorithms in terms of resolution and contrast, as calculated using the FRC metrics (Fig. 4 and Table 2).ConclusionsIn this study, a self-adaptive mixed-emitter single-molecule localization algorithm that enables the adaptive determination of the fitting area and fitting number is proposed. Compared to the SE and ME algorithms, the SM algorithm can significantly reduce the artifacts caused by mismatch and crosstalk errors, resulting in an enhanced resolution and contrast within the full applicable density range of the fitting method. In terms of the speed, the current SM algorithm is faster than ME algorithm by a factor of 3?4, and slower than the SE algorithm by one order of magnitude. However, the number of fitting iterations required by the SM algorithm is the same as that required by the SE algorithm. Therefore, after optimization, the SM algorithm has the potential to achieve a speed comparable to that of the SE algorithm. Although the analysis and experiments in this study were conducted under two-dimensional and single-channel conditions, the inherent mechanism of the SM method allows for its easy integration with more complex single-molecule imaging technologies, such as three-dimensional and multi-channel situations. In future research, the SM algorithm should be further refined and its reliability and stability should be verified, thereby expanding its advantages in the field of biological imaging.
SignificanceBiomedical ultrasound imaging has been widely used as an imaging technology based on ultrasound signals for viewing the internal structure of the body and finding the source of diseases. In recent decades, owing to the development of ultrasonic transducers, ultrasound imaging has made significant progress in obtaining important diagnostic information using rapid and noninvasive methods. Traditional transducers are excited by electricity and take advantage of the piezoelectric effect to achieve a transformation between electricity and ultrasound. However, considering the demanding requirements of application environments, the primary restriction is the limited bandwidth of traditional transducers.Laser-generated ultrasound, a novel technology based on photoacoustic effect, is excited by a laser instead of electricity. Ultrasound pulses are generated by the absorption of a short-pulse laser, thus leading to elastic thermal expansion caused by the transient temperature increase. In this process, the time-varying laser acts as the only excitation source. The upper limits of the energy and frequency of the ultrasound are restricted by the laser. Compared with piezoelectric transducers, the ultrasound generated by laser-generated ultrasound transducers has the characteristics of high frequency and large bandwidth, which are necessary for sensing and imaging.With the breakthrough of laser-generated ultrasound transducers in the structural simplification and excitation of large-bandwidth ultrasound, laser-generated ultrasound technology has been gradually applied in various fields where traditional piezoelectric ultrasound methods cannot be applied, essentially providing a novel idea for high-precision and high-resolution biomedical applications.ProgressThe amplitude of ultrasound produced by laser-generated ultrasound technology is related to various characteristics, such as laser energy, transducer absorbance, thermal expansion coefficient, and shape. Moderately high-energy laser, highly absorbing nano-scale light absorbers, and expanders with high thermal expansion coefficients positively affect the ultrasound amplitude generated by laser-generated ultrasound transducers. Meanwhile, the ultrasound frequency domain generated by photoconductive ultrasound technology is related to parameters such as the excitation light pulse width, transducer material, and transducer thickness (Figs. 2?4). For example, under test conditions in which the imaging depth is small but the imaging resolution is very high, an ultra-narrow pulse width laser with a nanoscale metal layer can be used as an optical ultrasound transducer (Table 1 and Fig. 2). If the test environment has high requirements for imaging depth and imaging speed but low requirements for imaging resolution, a common nanosecond transducer is suitable. If the test environment has high requirements for imaging depth and speed and low requirements for resolution imaging, a common nanosecond-pulsed laser with the carbon-based polymer material is suitable as a solution for ultrasound.Moreover, the less complicated structure of the laser-generated ultrasound transducer promises a large amplitude of the ultrasound at the focal point, with a self-focusing effect when using a concave transducer (Figs. 1 and 5). Furthermore, the ultrasound generated by a laser-generated ultrasound transducer has a high frequency and large bandwidth, thereby contributing to a smaller sound field at the focal point (Fig. 6).Conclusions and ProspectsThis study summarizes the mechanism of action, transducer system, performance characterization, and application areas of phototransduction ultrasound technology, as well as the applications of concave transducers in neural stimulation, ultrasonic cavitation, and ultrasound imaging, and describes the advantages and disadvantages of piezoelectric-based and photoacoustic effect-based transducers by comparing them with conventional ultrasound transducers. With the continuous development of theoretical systems of laser-generated ultrasound and precision processing technology, the advancement of laser-generated ultrasound technology has led to new opportunities for the development of biomedical ultrasound.
ObjectiveThe considerable potential of temperature-responsive nanomaterials as contrast agents has driven research and development in the field of photoacoustic imaging in recent years. However, the temperature-sensitive nanoprobes currently reported have response temperatures that exceed the tolerable range of humans, and their reversibility is low, which are two serious problems that hinder imaging and long-term monitoring in practical applications. Molecular photoacoustic imaging has emerged as a noninvasive imaging modality for cancer diagnosis, which couples superb optical absorption contrast and an excellent ultrasonic spatial resolution. However, research on molecular photoacoustic imaging has focused on optical wavelengths in the visible and near-infrared (NIR) part of the spectrum between 550 and 900 nm, with a relatively low sensitivity and limited imaging depth. Of note, the characteristic molecules of many major diseases—particularly in the early stage—exhibit no obvious photoacoustic contrast in the optical window of biological tissue (NIR-Ⅰ, 650?950 nm; NIR-Ⅱ, 950?1700 nm). Moreover, most of the current photoacoustic contrast agents are “always-on” probes, which can only provide invariable imaging contrast and struggle to eliminate the inherent background effect of biological tissues. In contrast, activable photoacoustic contrast agents can respond to a given cancer biomarker and emit signals. Therefore, there is an urgent need to develop a stimulus-responsive photoacoustic probe for the second NIR region. In this paper, a reversible temperature-responsive phase-change AuNR@PNIPAM nanoprobe is proposed that can dynamically modulate the temperature field through an external NIR optical switch to obtain contrast-enhanced photoacoustic images.MethodsWhen the temperature-sensitive AuNR@PNIPAM microgel is delivered to tumor tissue under 980 nm laser irradiation, the gold nanorod core absorbs NIR light energy, triggering a phase transition of the PNIPAM shell. As the temperature exceeds the volume phase transition temperature (VPTT) of PNIPAM, the PNIPAM hydrogel undergoes a sol-gel phase transition, which increases the refractive index around the gold nanorod, inducing a redshift of the localized surface plasmon resonance band and an increase in the absorption-peak intensity of the microgel. Therefore, these microgels exhibit enhanced and switchable NIR absorption in the physiological temperature range, allowing high-contrast imaging of tumors.Results and DiscussionsThe authors characterized the morphology and temperature response of the synthesized AuNR@PNIPAM microgel. It was observed that PNIPAM was successfully compounded onto AuNR, and when the temperature exceeded the VPTT of PNIPAM, the volume change of the PNIPAM hydrogel resulted in a change in the refractive index, enhancing the absorption in the second NIR region, as illustrated in Fig. 2. Furthermore, the microgel exhibited a high and stable photothermal conversion efficiency in in vitro and in vivo photothermal experiments, as shown in Fig. 4. The experimental results in Fig. 5 confirmed that the prepared AuNR@PNIPAM microgel could enhance the photoacoustic imaging contrast when an external NIR optical switch was used to trigger the temperature phase transition. Therefore, owing to their excellent photoacoustic imaging contrast ability, AuNR@PNIPAM microgels have considerable potential for early cancer diagnosis and hyperthermia detection.ConclusionsIn this study, the photoacoustic signal of AuNR@PNIPAM microgels was amplified near the physiological temperature, indicating their feasibility for high-contrast photoacoustic imaging of tumors. AuNR@PNIPAM microgels exhibited improved NIR-Ⅱ absorption under 980 nm laser irradiation, because of a redshift of the localized surface plasmon resonance band and an increase in the absorption-peak intensity. Furthermore, the prepared AuNR@PNIPAM microgels exhibited reversible temperature-responsive characteristics and an efficient and stable photothermal conversion effect; hence, they could modulate the temperature field through the NIR optical switch to realize the reversible switching of NIR-II absorption. Thus, these microgels could control the turning on/off of the photoacoustic signal to suppress the unwanted background signals. This work proposes a strategy for achieving high-contrast imaging of tumors by dynamically responding to temperature stimuli, providing guidance for the development of temperature-responsive smart photoacoustic probes for enhancing imaging contrast.
SignificanceOptical coherence tomography (OCT) is a label-free optical imaging technique based on the principle of low-coherence interference, which has the advantages of high resolution and fast imaging speed. OCT can image tissue anatomy and microcirculation without physiological sections and exogenous contrast agents. However, the OCT penetration depth is limited to 2?3 mm owing to the optical scattering of biological tissue. Therefore, most OCT applications focus on ocular imaging and endoscopy. OCT has led to a better understanding of ocular structures and has provided efficient treatment of glaucoma, maculopathy, and other ocular diseases. Endoscopy is another important field of OCT application. Combining OCT imaging with an endoscopic micro-probe, endoscopic OCT can obtain three-dimensional morphological microstructures of in vivo internal organs with depth-resolved information and micron-scale resolution, which is advantageous in detecting small lesions under the surface of tissue. With an optical fiber and a miniaturized lens, an endoscopic OCT probe can be inserted into the body through the working channel of a conventional video endoscope. By overcoming the low resolution of ultrasound imaging and the shallow penetration depth of confocal imaging, endoscopic OCT has become an indispensable imaging tool in clinical diagnosis.ProgressFirst, we summarize the development of endoscopic OCT over recent years. Although ophthalmic OCT still predominates, the research and application of endoscopic OCT techniques are increasing (Fig.1). Three types of OCT systems are described: time-domain OCT, spectral-domain OCT, and swept-source OCT (Fig.2). In contrast to time-domain OCT systems with the mechanical scanning structure in the reference optical path, frequency-domain OCT systems, including spectral-domain OCT and swept-source OCT, record the interference signals as functions of wavelength. The depth information of the sample can be obtained by the Fourier transform of the interference signals at different wavelengths. Frequency-domain OCT improves imaging acquisition speed. Then, various probes are presented, such as the anterior and side-view imaging probes (Figs.2 and 3). An anterior imaging probe with the beam along the optical axis is suitable for guiding surgical procedures. A side-view imaging probe is easily minimalized and is capable of imaging tissue with small inner diameters, such as blood vessels.Second, the various techniques of endoscopic OCT are summarized, including ultrahigh-resolution OCT and dual-modality imaging. Imaging of porcine coronary arteries with ultrahigh-resolution OCT can detect lesions in the endothelial cell layer, providing a new option for the early diagnosis of coronary atherosclerosis (Fig.5). The alveolar structure in human lung tissue can be observed clearly using ultrahigh-resolution OCT imaging (Fig.6). Ultrahigh-resolution OCT may have more applications in clinical practices if the cost can be deduced. Multimodality imaging has become a popular research area in recent years, which can acquire multiple images simultaneously and overcome the limitations of OCT, providing precision clinical diagnosis. Dual-modality imaging combining OCT with fluorescence imaging compensates for the lack of molecular sensitivity in OCT and provides more detailed information about the tissue (Fig.7). Dual-modality imaging with OCT and ultrasound combines the advantages of the high resolution of OCT and the deep penetration of ultrasound imaging to acquire two types of structural information simultaneously (Fig.8).Third, we introduce commercialized endoscopic OCT and compare the performance. Many endoscopic OCT devices have emerged over recent years (Table 2). From the clinical applications of endoscopic OCT technology, we review the current advances in cardiology, respirology, gastroenterology, urology, and gynecology. In cardiology, OCT applications for atherosclerosis assessment (Fig.9) and postoperative evaluation of stent implantation procedures have been introduced (Fig.10). In respirology, OCT endoscopy technology has increasing applications in the early diagnosis of lung cancer, chronic bronchial inflammation, bronchial asthma (Fig.12), etc. In gastroenterology, OCT endoscopy can diagnose Barrett’s esophagus lesions early with the risk of esophageal adenocarcinoma (Fig.13). Although endoscopy imaging is challenging for intestinal tissue owing to the large size of the stomach and the long length of the small intestine, endoscopic OCT has promising applications in areas such as intestinal damage diagnosis (Fig. 14) and small intestine allografts (Fig.15). In addition, endoscopic OCT has been used for cancer screening of various tissues, such as the biliopancreatic duct, cervix, and ureter, providing an accurate diagnosis of neoplastic lesions (Fig.17). In gynecology, endoscopic OCT technology offers new ideas for diagnosing gynecological diseases and monitoring of vaginal health status (Fig. 18).Conclusions and ProspectsEndoscopic OCT technology has progressed from time-domain OCT to frequency-domain OCT in the past few decades and has become an essential diagnostic tool in addition to traditional endoscopic imaging methods. However, endoscopic OCT technology still requires continuous improvement, including the enhancement of imaging quality, the miniaturization of probes, the extension of imaging depth, the improvement of spatial resolution, reduction in manufacturing costs, and combination with other imaging modalities. With an improvement in performance, endoscopic OCT technology will provide a more significant imaging basis for precision medicine.
SignificanceEukaryotic cells have numerous cellular structures, including a variety of organelles and macromolecular complexes. These structures have specific physiological functions and work interactively to perform certain cellular activities. Therefore, studying these structures in their native state is essential to understand the real physiological processes in the cells. In situ investigation of cellular structures does not only provide morphology, distribution, and abundance information, but also reveals their interaction mechanisms, thereby providing new insights into the understanding of life.Cryo-electron tomography (cryo-ET) is currently the principal technique to resolve the in situ structures of biological specimens. By collecting tilted series of transmission electron images and performing image reconstruction, cryo-ET determines the 3D structures of bio-specimens with a nanometer-level resolution. A prerequisite for applying cryo-ET is to fix the sample under cryogenic conditions. High-pressure freezing and plunge freezing are well-established cryo-fixation methods that preserve biological specimens in their near-native state in vitreous ice. Benefiting from these techniques, cryo-ET has been widely applied to cells and tissues.One limitation to cryo-ET is its restricted imaging depth, which is typically a few hundred nanometers owing to the confined penetration capabilities of electrons. Therefore, reducing the thickness of the samples to that of lamellae of approximately 200 nm is necessary before applying cryo-ET. Focused ion beam (FIB) milling has been recently employed to prepare lamellae of bio-specimens for cryo-ET. Compared to traditional ultramicrotomy, FIB milling avoids artifacts such as distortions, crevasses, and compression when fabricating the lamella. However, conventional FIB milling does not allow site-specific milling, because in a dual-beam FIB/SEM system, FIB or SEM image only illustrates the surface morphology of the sample and cannot provide more information to recognize and localize the underlying interest targets. When milling cells with FIB, cutting at an arbitrary position can only hit abundant cellular structures such as Golgi apparatus or mitochondria but cannot be used to prepare lamellae containing specific targets. This drawback hinders the application of FIB in cryo-ET.The “blind” milling can be improved by correlative light and electron microscopy (CLEM). In CLEM, the targets of interest are fluorescently labelled and can be identified by fluorescence imaging. After registering the light and FIB images, fluorescence signal can be used to guide the FIB to mill at specific sites. Currently, various light imaging modalities have been adopted to navigate FIB fabrication, including widefield microscopy, confocal microscopy, and Airyscan. Moreover, two major working routines, that is, pipelined and integrated workflows, have been established to perform fluorescence-guided FIB milling. Therefore, it is important and necessary to summarize the existing techniques and discuss the advantages and limitations of different working routines to provide guidelines for researchers to choose the appropriate protocols.ProgressThis study reviews the essential techniques involved in fluorescence-guided cryo-FIB milling. First, plunge freezing is introduced. Plunge freezing is the most commonly used technique to vitrify cells. The key aspects to obtain good plunge-frozen specimens are discussed, including the choice of electron microscope grids and supporting films (Fig. 2), available commercial instruments (Fig. 3), and standard protocols.Second, as a popular method to prepare lamellae of vitrified cells, FIB milling is discussed in several aspects: the working principle is introduced; the relevant instrumentations are summarized, including dual-beam FIB/SEM system (Fig. 4), cryostage and cryotransfer systems (Fig. 5), and Autogrid and sample holder (Fig. 6); and the milling of frozen cells is outlined (Fig. 7).Third, the principle (Fig. 8) and workflow (Fig. 9) of fluorescence-guided FIB milling is introduced. Pipelined and integrated workflows are described, and relevant commercial instruments are overviewed (Figs. 10 and 11). The different workflows and various systems are compared (Table. 1). The most recent developments of integrated solutions are discussed in detail. Sun Fei's research group and Ji Wei's research group from the Institute of Biophysics, Chinese Academy of Sciences have developed novel integrated light, ion, and electron microscopies (Figs. 12 and 13), thereby providing new avenues for performing accurate and efficient FIB milling at specific sites under fluorescence guidance.Conclusions and ProspectsIn situ investigation of cellular structures using cryo-ET has recently become an interesting research topic. Fluorescence-guided FIB milling has been applied to mill vitrified biological samples at specific sites. The recent developments in integrated cryo-FLM-FIB/SEM systems and workflows provide efficient and accurate methods to fabricate cell lamellae containing desired targets. These innovations have the potential to serve as all-in-one solutions for site-specific cryo-lamella preparation for cryo-ET in the future.
Significance Various biological imaging modalities have become essential tools in life science research, preclinical research, and clinical practice. The emergence of enormous in vivo imaging technologies such as computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET), and single-beam emission computed tomography (SPECT) plays a significant role in disease diagnosis, progression monitoring, and prognosis, bringing the possibility of molecular imaging into medical observation.Although they have unlimited penetration depth, the above techniques suffer from disadvantages such as limited spatial resolution, long operation times, and low sensitivity. Additionally, the equipment is often very expensive and induces radiation. On the other hand, as a radiation-free technique, fluorescence imaging has been widely used for in vivo imaging due to its high spatiotemporal resolution and labeling specificity. However, the performance of FL imaging is deteriorated by the strong absorption, scattering, and autofluorescence of biological tissues in the visible (400?700 nm) or NIR-I (700?900 nm) regions and shows unsatisfactory penetration depth, spatial resolution and signal-to-noise ratio (SNR), limiting its further application in in vivo imaging.FL in the second near-infrared region (NIR-II, 1000?1700 nm), on the contrary, shows appealing advantages due to its deeper penetration depth (>10 mm), improved spatial resolution (about 3 μm), and higher signal-to-noise ratio (about 20), unveiling great clinical translation. Since single-walled carbon nanotubes (SWNTs) were first applied to NIR-II fluorescence imaging in small animals, the development of NIR-II fluorescent probes with high molar absorbance coefficients, high fluorescence quantum yields, good stability, and good biocompatibility has been a research hotspot. In the past decade, NIR-II fluorescent probes were primarily classified into two categories: organic and inorganic probes. The NIR-II inorganic fluorescent probes (e.g., single-walled carbon nanotubes, quantum dots, and rare earth doped conversion materials) have strong heavy metal toxicity, are typically poor in biocompatibility, and have difficulty completing physiological metabolism in vivo, limiting their potential applications in clinical practice. On the other side, organic NIR-II fluorescent probes are free of heavy metal ions and have clear structures as well as better biocompatibility, making them more suitable and promising for clinical translation. Some representative examples are D-A-D small molecules, cyanine dyes, and conjugated polymers. Hence, to guide the future development of this field more rationally, it is important and necessary to summarize the molecular structure design concepts and biomedical imaging applications of organic NIR-II fluorescent probes.ProgressIn this review, we systematically summarize the molecular structure design concepts and biomedical imaging applications of organic NIR-II fluorescent probes reported in the current literatures. The research progress of organic NIR-II fluorescent probes is classified into anatase dyes, D-A-D organic small molecules, and conjugated polymers.First, the molecular design strategies of cyanine dyes with NIR-II emission wavelengths are summarized in terms of red-shifting absorption/emission wavelength, improving fluorescence quantum yield, enhancing biocompatibility, and chemical stability, respectively. Up to now, the reasonable and result-oriented design strategies to achieve cyanine dyes with NIR-II emission wavelengths primarily include: 1) extending the effective conjugation system, 2) modifying the donor and acceptor units, and 3) constructing fluorophore J-polymer. The strategies to enhance the fluorescence brightness primarily include: 1) introducing spatial site resistance, 2) forming complexes with proteins, and 3) enhancing the rigidity of molecular structures. The effective strategies to improve biocompatibility primarily include: 1) encapsulating hydrophobic fluorescent molecules by nanoprecipitation using amphiphilic materials and 2) introducing hydrophilic groups on hydrophobic fluorescent molecules utilizing molecular engineering.Second, the development process of D-A-D small molecules in terms of donor/acceptor unit modulation and fluorescence quantum efficiency enhancement is also presented. In 2016, Dai's team reported for the first time that the water-soluble small molecule CH1055-PEG could be used for NIR-II fluorescence imaging. Since then, a series of small molecules with NIR-II emission have been designed by modulating the electron-giving/absorbing ability of donor/acceptor units. Moreover, strategies have been proposed to enhance fluorescence quantum efficiency, such as by introducing shielding units, suppressing TICT states, constructing hydrophobic nonpolar environments, and building fluorescent small molecules with AIE properties.Subsequently, we summarize the molecular design strategies of organic conjugated polymers with high brightness and further discuss their applications in bioimaging, primarily including tumor imaging, dynamic angiography, and photothermal therapy.Finally, the issues and challenges that need to be addressed to identify the clinical translation of NIR-II fluorescence imaging techniques are discussed.Conclusions and ProspectsNIR-II fluorescence imaging has been widely used in basic scientific research and preclinical practice. Organic NIR-II fluorescent probes are highly amenable to clinical translation due to their excellent biocompatibility, good synthetic reproducibility, and extremely high chemical modifiability. To date, a series of NIR-II fluorescent probes with excellent performance have been developed and applied for in vivo imaging with a high signal-to-noise ratio, deep-tissue penetrating ability, and high spatial and temporal resolution. However, most organic NIR-II fluorescent probes reported in the literatures are not yet well established and have limitations in clinical applications. To expand the biological applications of NIR-II fluorescent probes and to achieve true clinical translation, the following challenges must be overcome: 1) the development of liver/kidney metabolizable probes to address long-term probe safety; 2) the development of endogenous NIR-II fluorescent proteins for long-term biomonitoring; and 3) the development and optimization of NIR-II fluorescent imaging systems.
ObjectiveOptical coherence tomography (OCT) is widely employed for ophthalmic imaging and diagnosis because of its low latency, noncontact nature, noninvasiveness, high resolution, and high sensitivity. However, two major issues have hindered the development of OCT diagnostics for ophthalmology. First, OCT images are inevitably corrupted by scattering noise owing to the low-coherence interferometric imaging modality, which severely degrades the quality of OCT images. Second, low sampling rates are often used to accelerate the acquisition process and reduce the impact of unconscious motion in clinical practice. This practice leads to a reduction in the resolution of OCT images. With the development of deep learning, the use of neural networks to achieve super-resolution reconstruction of OCT images has compensated for the shortcomings of traditional methods and has gradually become mainstream. Most current mainstream super-resolution OCT image reconstruction networks adopt convolutional neural networks, which mainly use local feature extraction to recover low-resolution OCT images. However, traditional models based on convolutional neural networks typically encounter two fundamental problems that originate from the underlying convolutional layers. First, the interaction between the image and convolutional kernel is content-independent, and second, using the same convolutional kernel to recover different image regions may not be the best choice. This often leads to problems, such as excessive image smoothing, missing edge structures, and failure to reliably reconstruct pathological structures. In addition, acquiring real OCT images affects the training effectiveness of previous models. First, deep learning models usually require a large amount of training data to avoid overfitting; however, it is difficult to obtain a large number of real OCT images. Second, even if the results are excellent, it is meaningless to train the model without using images acquired from OCT devices commonly used in today’s clinics. To address the above problems, this study proposes a new OCT image super-resolution model that has the advantages of a convolutional neural network and incorporates a transformer to compensate for its disadvantages, while simultaneously solving the data aspect problem considering recent real clinical images and data enhancement methods during training to increase the generalizability of the model.MethodsIn this study, a transformer-based TESR for OCT image super-resolution network was constructed. It constituting three parts: a shallow feature extraction module, a deep feature extraction module, and an image reconstruction module. First, the input image is fused with the extracted edge details using the edge enhancement module, and then shallow feature extraction is performed using a basic 3×3 convolution block. The deep feature extraction module comprises six feature fusion modules, FIB, and a convolution block to extract more abstract semantic information. The FIB module comprises six newly proposed pyramidal long-range transformer layers, PLT, and a convolutional block. The PLT module fuses two mechanisms of local and global information acquisition, where the shifted convolutional extraction module is used to expand the perceptual field and effectively extract local features of the image, and the pyramidal pooling self-attention module is used to strengthen the attentional relationships between different parts of the image and capture feature dependencies over long distances. Finally, image reconstruction was completed using a pixel-blending module.Results and DiscussionsWe compare our model with four classical super-resolution reconstruction models for 2× and 4× reconstruction, namely, SRGAN, RCAN, IPT, and SwinIR. Quantitative evaluation metrics include the peak signal-to-noise ratio (PSNR), structural similarity (SSIM), and learning perceptual image patch similarity (LPIPS). For qualitative evaluation, we provide 4× reconstructed images sampled from both datasets for comparison. The experimental results show that TESR outperformed the other methods on both datasets. Objectively, the PSNR results of TESR improved by 7.1%, 6.5%, 3.2%, and 1.9%, the SSIM results improved by 5.9%, 5.3%, 3.5%, and 2.2% (Table 1), and the LPIPS results decreased by 0.1, 0.13, 0.06, and 0.01 (Table 2) for the 4× image reconstruction. Similar results are obtained for 2× image reconstruction. Zooming in on the key reconstructed areas, it is clear that the TESR-reconstructed images can better restore the hierarchical information of the retina using the edge enhancement module and image feature extraction (Fig. 9). The retinal edge structure is sharp, the texture details are clear, and there are no obvious noise or artifact problems (Fig. 10). The overall image is clean with high realism and is close to the HR reference image. The experiment verifies the effectiveness and superiority of TESR for super-resolution reconstruction of OCT images.ConclusionsTo address the problems that OCT image super-resolution reconstruction algorithms focus too much on local features and ignore the internal knowledge of the overall image, while lacking the extraction of retinal edge details, we proposes a transformer-based edge enhancement OCT image super-resolution network TESR. TESR restores the edge detail information of OCT images with high quality through the new edge enhancement module, while suppressing the noise problem of the images. The PLT module used in deep feature extraction further fuses the local and global information of the image to model the overall internal information of the image over a long range. This approach eliminates the artifact problem that tended to occur in previous algorithms and improves the realism of the reconstructed images. The experiment shows that the TESR model proposed in this study is better than other classical methods in terms of PSNR and SSIM, respectively. It is excellent in terms of LPIPS, and has a significant improvement in subjective visual quality. Additionally, the model has a strong generalization ability. In the future, more effective self-attentive implementations will be explored to reduce the computational complexity of the transformer and improve the convenience of the super-resolution reconstruction technique for clinical practice.
ObjectiveCherenkov-excited luminescence scanning imaging (CELSI) is an emerging optical imaging technology that provides a new tool for tumor diagnosis and treatment. However, CELSI image reconstruction is ill-posed and underdetermined because of light scattering in biological tissues and limited boundary measurements. Regularization techniques have been widely adopted to alleviate the ill-posedness of the CELSI reconstruction. However, these methods typically exhibit poor image quality. To date, deep-learning-based reconstruction algorithms have attracted significant attention in optical tomography. To enhance the image quality of CELSI, we develop a reliable and effective deep learning reconstruction algorithm based on unrolled iterative optimization.MethodsIn this paper, a deep learning reconstruction algorithm is introduced based on unrolled iterative optimization, which takes the acquired sinogram image as network input and directly outputs the high-quality reconstructed images through end-to-end training. First, the image reconstruction of CELSI is reformulated as a l1 norm optimization problem based on sparse regularization technique. Second, the alternating direction method of multipliers (ADMM) based neural network algorithm (ADMM-Net) is adopted to minimize the optimization problem, which converts each iteration into convolution neural network (CNN) processing layer and deploys multiple processing layers cascaded into a deep network. Each processing layer consists of a reconstruction layer, a nonlinear layer, and a multiplier update layer. We linearize the reconstruction layer to avoid matrix inversion. The nonlinear transformation function in the nonlinear layer consists of five convolutional operators with three rectified linear unit (ReLU). The first convolution operator comprises 32 filters with the size of 3×3, and the other convolution operators consist of 32 filters with the size of 3×3×32. Note that all the parameters in the ADMM-Net are end-to-end updated through gradient backpropagation, including the step size and regularization parameter in the reconstruction layer, the shrinkage threshold in the nonlinear layer, as well as the nonlinear transformation function. To evaluate the effectiveness of ADMM-Net, numerical simulation experiments were performed, and the performance was compared with the traditional FBP algorithm, two deep learning algorithms (FBPConvNet and ISTA-Net+). Root mean square error (RMSE), peak signal-to-noise ratio (PSNR), and structural similarity (SSIM) are used as quantitative metrics.Results and DiscussionsFirst, the influence of the number of layers on the reconstruction performance is evaluated. Our results show that a better image quality can be obtained when the number of layers increases (Table 1). However, a large number of layers increases the computational cost. To balance the quality of the reconstructed images and computational cost, the number of layers is chosen empirically as five. Furthermore, experiments are performed using a single fluorescent target. Compared with the other three algorithms, reconstructed images with fewer artifacts are obtained for the proposed ADMM-Net (Fig.5). The quantitative results show that the average values of PSNR and SSIM yielded by our algorithm are 33.75 dB and 0.86, respectively, and compared with the other three algorithms, ADMM-Net achieves the average reduction of 81.5%, 16.3%, and 25.2% in RMSE, improvement of 77.8%, 5.1%, and 8.6% in PSNR, and increases of 47.2%, 4.5%, and 2.7% in SSIM (Table 2). The ability of ADMM-Net to distinguish between two targets is also tested. When the edge-to-edge distance between the two fluorescent targets is 2 mm, the FBP, FBPConvNet, and ISTA-Net+ algorithms fail to separate the two fluorescent targets, whereas ADMM-Net successfully distinguishes the two fluorescent targets (Fig.8). The plot of the profiles again confirmed that ADMM-Net can achieve satisfactory results in terms of the reconstructed positions, sizes, and quantum yields of the fluorescent targets (Fig.9). Finally, the generalizability of ADMM-Net is verified by reconstructing three and four fluorescent targets with different ratios of fluorescence quantum yields. Our results reveal that the FBP, FBPConvNet, and ISTA-Net+ algorithms cannot accurately recover the distribution of fluorescent targets. In contrast, the proposed algorithm exhibits a good generalization performance and can accurately recover the distributions of three and four targets (Fig.11).ConclusionsThis paper proposes a deep learning reconstruction algorithm for CELSI based on unrolled iterative optimization (ADMM-Net), which combines the structure of the iterative update steps of the ADMM algorithm with a CNN. The performance is evaluated through numerical simulation experiments. As a result, compared with the FBP, FBPConvNet, and ISTA-Net+ algorithms, the proposed algorithm can yield better results with fewer artifacts and higher quantitative accuracy when reconstructing single fluorescent targets with different contrasts. Moreover, the proposed algorithm can distinguish between two fluorescent targets with high image quality, even when the edge-to-edge distance is 2 mm. Our results also demonstrate that our algorithm has a good generalization performance and can reconstruct three and four fluorescent targets accurately, even when the model is trained on a dataset with only one or two targets.
ObjectiveHepatocellular carcinoma (HCC) is the most common primary liver malignancy and the second leading cause of cancer death worldwide. The development of HCC leads to abnormalities in the structure and function of blood vessels, which further lead to high pressure and hypoxia in the tumor microenvironment (TME). The most common clinical methods for identifying HCC nowadays are magnetic resonance imaging (MRI), computed tomography (CT), and ultrasound. MRI can measure multifunctional parameters of the liver; however, it has significant limitations in imaging resolution and is costly. CT can image the blood vessels in the liver; however, it uses X-ray, thus increases the risk of cancer. Ultrasound imaging is widely used to evaluate HCC; however, its sensitivity and specificity are low. Therefore, a more complete and reliable technique to analyze the micro-vascular morphology of HCC and TME is urgently needed. Photoacoustic imaging is a rapidly developing imaging technology in recent years. It offers a wide range of potential applications in the field of medical imaging and can visualize the structure and function information of biological tissues without labeling of contrast agents or invasion. Photoacoustic imaging has high specificity and sensitivity in the diagnosis of HCC and can visualize functional imaging of tumors and morphological examination of blood vessels.MethodsA mouse model of in situ liver cancer was established, and the bioluminescence signal was activated by an in vivo fluorescence imaging system to locate the tumor. The microvascular structure characteristics and oxygen saturation of normal liver lobules, tumor centers, and adjacent tumors were accurately observed using photoacoustic microscopy. The concentrations of oxygenated and deoxygenated hemoglobins were quantified using the spectroscopic separation method to calculate blood oxygen saturation. The photoacoustic images were converted into binary images, and the vascular signals were extracted for density and diameter analysis.Results and DiscussionsThe results obtained using photoacoustic microscopy via two wavelengths (532 nm/559 nm) show that the blood vessels in the normal liver are evenly spaced and well differentiated, whereas large irregular vessels appear at the edges of the tumors, and the vascular joints are curved and dilated. The blood vessels inside the tumors are unevenly distributed and the branch diameter increases. The oxygen concentration in the blood around the tumors decreases, resulting in a hypoxic and high-pressure TME.ConclusionsIn microvascular monitoring of hepatocellular carcinoma, the photoacoustic imaging can provide high-resolution images, which can more accurately detect the morphology of tiny and abnormal blood vessels, improving the accuracy of early cancer detection. Through image analysis, indicators such as the density and diameter of microvessels and other information such as oxygenation level and metabolic activity of tumor tissues can be evaluated to assess the growth state of tumors and predict the degree of malignancy. Photoacoustic imaging demonstrates a high application potential for studying the development of HCC. It can provide further insights into the antiangiogenic therapy of tumors and the diagnosis of numerous liver-related diseases.
ObjectiveA human mature erythrocyte membrane skeleton is a triangular lattice network composed of various proteins under the membrane, which is essential for the maintenance of cell morphology, deformation, movement, and metabolism. The unique ultrastructural arrangement of the erythrocyte membrane skeleton is fascinating and has attracted many scientists to develop new technologies for imaging and analysis. Emerging single-molecule localization super-resolution microscopy (SMLM) has demonstrated significant capability in resolving the nanoscale ultrastructure of the erythrocyte membrane skeleton; however, the improvement of resolution has put forward high requirements for imaging analysis methods. A Vorono? diagram is a geometric analysis method that divides points in space into different regions to describe their spatial distribution. It is widely used in space exploration, materials science, machine learning, and other research fields. In recent years, this method has been prominently utilized in SMLM data extraction and analysis, mainly in the clustering and colocalization analysis of “point cluster”-shaped images. Taking advantage of the Vorono? method particularly in SMLM image analysis, we aim to apply this method to extract the distribution information of erythrocyte membrane skeleton protein SMLM images, to more quantitatively and accurately reveal skeletal organization characteristics.MethodsSMLM super-resolution images of erythrocyte membranes and skeletal proteins were obtained using a self-built SMLM imaging system. Actin was stained with fluorescently labeled phalloidin (Alexa 647-phalloidin). CD59, N terminus of β-spectrin, tropomodulin (TMOD), and ankyrin were labeled with specific antibodies. After SMLM imaging, regions of interest in the SMLM images were selected for analysis, and the corresponding point-cloud image was drawn according to the positioning coordinates. The centroid of each point cluster was subsequently acquired using DBCAN clustering analysis, and the image boundary was determined based on the maximum and minimum values of all centroid coordinates. The obtained centroids were used as seed points for Vorono? tessellation, and the vertex coordinates of the Vorono? polygon generated by each seed point were obtained using the voronoin function in MATLAB. Area A of the Vorono? polygon was calculated using the polyarea function in MATLAB. All areas A were divided by the average area〈A〉to obtain a histogram of the area distribution (Fig. 1). Finally, the area distribution of the Vorono? polygon was fitted with the γ function, which could be used to describe the spatial distribution characteristics of the “point cluster”-shaped SMLM images of erythrocyte membrane and skeleton proteins.Results and DiscussionsFirst, Vorono? analysis was performed for CD59, an erythrocyte membrane protein with high lateral mobility. The x-axis corresponding to the peak of the γ distribution profile (xpeak) of CD59 was 0.78 (Fig. 2), which was slightly larger than the xpeak of the simulated points with a random distribution (Fig. 3). Considering the radius of the point-spread function in the SMLM imaging system, each simulated point was adjusted to a disk with a certain radius (set to 15 nm) for analysis. It was identified that the xpeak derived from the γ distribution of the normalized area increased with point density, while fluctuating between 0.78 and 0.8 in the density range of 60~100 μm-2, which was consistent with the CD59 Vorono? analysis result, indicating a random distribution of CD59 (Fig. 3). Furthermore, the xpeak values of the membrane skeleton proteins localized at the nodes of the skeleton triangular lattice network of the erythrocyte membrane, including actin, the N terminus of β-spectrin, and tropomyosin, were all 0.86, while the xpeak value of ankyrin was 0.84, indicating that these skeleton membrane proteins were distributed relatively uniformly, whereas the distribution of ankyrin was more random than that of other skeleton proteins (Fig. 4). To investigate the effects of deletion and disturbance of an erythrocyte triangular lattice skeleton on Vorono? analysis results, a Vorono? tessellation of simulated points was conducted with a density considerable to that of actin (approximately 80 μm-2 measured by SMLM imaging) using a custom-written MATLAB routine. After generating simulated points with an 80 nm interval periodic triangular lattice distribution, random disturbances of varying degrees (0-0.5) relative to the lattice length were applied to the locations of all points, and some points were randomly removed such that the density was identical to that of actin (Fig. 6). The variation trend of xpeak was explored under different disturbance rates, and the results showed that xpeak was 0.86 when the disturbance rate was 0.15 (i.e., the skeleton disturbance was approximately 15%), which was consistent with experimental results, indicating that there was a disturbance of approximately 15% relative to the lattice length in the human erythrocyte triangle lattice skeleton (Fig. 6).ConclusionsIn this study, a solution based on a Vorono? diagram was proposed for the analysis of SMLM super-resolution images of the erythrocyte membrane skeleton. According to the SMLM images “point cluster” feature of membrane and skeleton proteins, we extracted the centroids of point clusters for Vorono? polygon tessellation, and introduced parameters including the x-axis coordinate xpeak corresponding to the peak value of Vorono? polygon area γ distribution curve, the variation coefficient Cv of the Vorono? polygon, and the peak value of the nearest distance for quantitative analysis and characterization of the spatial distribution of erythrocyte membrane and skeleton proteins. The results demonstrated that the accepted mobile membrane protein CD59 was randomly distributed on the cell membrane. Skeleton proteins that were considered to be localized at the triangular lattice nodes, such as actin, the N terminus of β-spectrin, and TMOD, showed a relatively uniform distribution with a disturbance rate of approximately 0.15, whereas the distribution of ankyrins on the spectrin skeleton was slightly less uniform than that on the lattice node. These results demonstrated the validity of the Vorono? method in evaluating the distribution characteristics of erythrocyte membrane skeleton proteins, and the method can be extended to extract and analyze information for other “point cluster”-shaped SMLM images. Finally, the Vorono? analysis strategy is beneficial for understanding accurate spatial distribution characteristics of membrane skeleton proteins and provides novel insights and methods for in-depth information extraction from SMLM super-resolution data.
ObjectiveThe multifocal structure light microscope (MSIM) can achieve an imaging depth of 50 μm and a diffraction limited resolution that is two times that of the traditional wide-field structure light technique. However, factors such as fluorescence noise, scattering, and aberration limit its chromatographic capability and imaging depth when conducting super-resolution imaging of thick samples. To overcome these limitations, the MSIM technique combined with two-photon excitation, is implemented to obtain a two-photon multifocus structured light illumination microscope (2P-MSIM), and the imaging depth and resolution of MSIM are thus improved. The MSIM has great application potential in the field of fast super-resolution microscopic imaging of deep tissues in vivo. However, owing to the diffraction limit, the enhancement of the existing 2P-MSIM spatial resolution is limited. To further improve the spatial resolution of 2P-MSIM, a two-photon sub-diffraction multifocal structure illumination microscopy (2P-sMSIM) is proposed in this paper.MethodsUsing the improved Gerchberg-Saxton (GS) phase recovery algorithm, the conditions restricting the amplitude and the phase were enhanced in specific locations on the spectrum plane (the input plane reached the output plane after Fourier transform); subsequently, the amplitude and phase were constrained. After several iterations, the phase diagram of the sub-diffraction spot arrays (SSAs) was calculated, and the initial SSAs were obtained. The sidelobe intensity was controlled at approximately one-tenth of the center intensity (the sidelobe effect can be eliminated by the subsequent image-processing algorithm), and the algorithm parameter optimization was completed to obtain an SSA suitable for system imaging. Raw data for SSA and normal diffraction limited spot arrays were obtained. Next, Gaussian pinhole filtering, pixel relocation, and deconvolution algorithms were used to obtain the SSA and normal diffraction limited spot array images. The resolution of 2P-MSIM imaging was improved under the excitation of the SSA.Results and DiscussionsThe simulation results for the SSA and normal diffraction limited spot arrays are shown in Fig. 4. The average value of the full width at half maximum (FWHM) of the normal diffraction limited spot arrays is approximately 10.60 pixel; the average FWHM for a single focal point in the SSA is approximately 7.64 pixel. As a result, the SSA is 72% of the size of the normal diffraction limited spot arrays, achieving its design purpose. The phase diagram designed above is loaded onto a liquid-crystal spatial light modulator, and the SSA generated by the 2P-sMSIM system is used to excite the uniform eosin solution. Normal diffraction limited spot arrays and SSAs are obtained, and the experimental results are shown in Fig. 6. The normal diffraction limited spot arrays have an average FWHM of 0.71 μm±0.07 μm for a single point, while the SSA has an average FWHM of 0.57 μm±0.07 μm. The SSAs are 80% the size of the normal diffraction limited spot arrays. SSAs can effectively reduce the size of the point diffusion function of the system to further improve the resolution of the 2P-MSIM system. To test the 2P-sMSIM spatial resolution, cell microtubules were used as samples for the experiments, and the results are shown in Fig. 7. the normal diffraction limited spot arrays have a resolution of 151 nm±5 nm after scanning and 135 nm±5 nm after SSA scanning. The image resolution improved by approximately 16 nm after the sub-diffraction-focusing lattice scanning. The experiments prove that the SSA designed using this algorithm can effectively improve the resolution of 2P-MSIM imaging. To further demonstrate the imaging effects of the 2P-sMSIM, commercial mitochondrial sections obtained from Invitrogen were used for fluorescence imaging. The results are presented in Fig. 8. The images after the SSA scan processing are clearer, and the image quality is better. A ring structure that cannot be distinguished in the normal diffraction limited spot array scanned images is clearly observed in Fig. 8(f). The experiments further prove that SSA can effectively improve the resolution and imaging quality of the 2P-MSIM system.ConclusionsA design method for SSAs is developed in this study and applied to a 2P-MSIM for experimental verification in bioimaging. In the simulation experiment, normal diffraction limited spot arrays are compared with the SSA. By calculating their standard deviations, the SSA can shrink the spots to 72% of the normal diffraction limited spot arrays. The SSA is reduced to 80% of that of the normal diffraction limited spot arrays, proving that the SSA can improve the spatial resolution. Compared with the normal diffraction limited spot arrays, microtubule imaging experiments further verify the effectiveness of the algorithm, and its resolution is improved by approximately 16-135 nm. Mitochondrial imaging experiments also demonstrate the effectiveness and practicability of this algorithm, which lays the foundation for further applications of this algorithm in super-resolution imaging of living cells and tissues.
ObjectiveAging is a major independent risk factor for aortic stiffness and cardiovascular diseases. The strength of the aorta is imparted by collagen fibers, which are the dominant fibrins within the aortic wall. Therefore, a three-dimensional (3D) quantitative assessment of age-related changes in the collagen fibers within the aortic wall is expected to provide important clues for research on cardiovascular diseases. Second harmonic generation (SHG) microscopy is an ideal tool for observing collagen fibers in biological tissues. Compared to the traditional histological analysis method, which requires tissue sectioning and staining, SHG microscopy has an intrinsic optical sectioning ability for the 3D imaging of intact tissues and allows label-free and high-specificity imaging of collagen fibers owing to its inversion-asymmetric and spatially ordered structure. Moreover, the high resolution, large depth penetration, low photobleaching and phototoxicity of SHG microscopy have significantly benefited the detailed imaging of thick tissues such as the aortic wall. However, the SHG-based 3D quantitative assessment of aortic collagen fibers has not yet been extensively demonstrated in aging-related research. In this study, we proposed combining SHG imaging with a representative spatial texture analysis algorithm, a 3D gray-level co-occurrence matrix (GLCM), to investigate age-related changes in the aorta from the perspective of collagen fiber microstructures. We hope that the proposed method and our findings can provide novel strategies and potential indicators for aortic aging assessment, and further benefit studies on age-related cardiovascular diseases.MethodsWistar-Kyoto (WKY) rats at 3 weeks (3 w), 12 weeks (12 w), and 44 weeks (44 w) were used in this study. First, the abdominal aortas were removed, cleaned, and cut open along the longitudinal axis. Subsequently, en-face 3D SHG imaging of the inner and outer surfaces of the aortic wall was performed using a commercial multiphoton microscope (A1R-MP; Nikon). Then, 11 texture feature parameters, including the correlation, contrast, entropy, energy, sum mean, variance, homogeneity, cluster shade, cluster prominence, max probability, and inverse variance, of the aortic collagen fibers were extracted from the 3D SHG image stacks using the 3D GLCM algorithm (Fig. 1). Finally, statistical analysis based on one-way ANOVA and Tukey's multiple comparison test was performed using GraphPad Prism software to sift out aging-associated features.Results and DiscussionsBy comparing the SHG images of the WKY rats of different ages, we found that the aortic collagen fibers gradually became thicker, less dense, and more evenly distributed from 3 w to 12 w and 44 w (Figs. 2 and 3), regardless of the intima, media, or adventitia. However, the general morphology of the collagen fibers in the aortic intima and media was remarkably different from that in the aortic adventitia. The intima and media collagen fibers were relatively straight (Fig. 2), whereas the adventitial collagen fibers were arranged in curved bundles and had stronger SHG signals (Fig. 3). The 3D GLCM analysis and statistics of the aforementioned SHG images further showed that in the aortic intima and media, six texture features of the collagen fibers, including the correlation, contrast, entropy, sum mean, variance, and homogeneity, were significantly different among the three age groups. These features characterized the consistency, clarity, strength heterogeneity, overall strength, strength concentration, and structural isotropy of the fiber textures (Fig. 4). Similarly, for the adventitial layers, three aging-associated textural features-the sum mean, variance, and homogeneity-were sifted out (Fig. 5). The age-related changes revealed by these preferential texture features were generally consistent with those observed in the 3D SHG image stacks. These results demonstrated that combining SHG imaging with the 3D GLCM algorithm is a practical strategy for assessing aging-related changes in the collagen fibers in the aortic wall, and that 3D GLCM texture features such as the correlation, contrast, entropy, sum mean, variance, and homogeneity are promising quantitative indicators of aorta aging.ConclusionsThis study proposed a novel strategy that combined SHG imaging with 3D GLCM for aortic-aging assessment from the fresh perspective of the collagen fiber microstructure. The collagen fibers within the aortic intima-media and adventitia of WKY rats with different weeks of age were imaged using SHG microscopy. The 3D GLCM was then used to quantify the stere omicrostructural characteristics of the collagen fibers based on the 3D SHG image stacks, and a variety of aging-related texture features, including the correlation, contrast, entropy, sum mean, variance, and homogeneity, were sifted out. The proposed method and derived texture features are expected to provide a powerful tool and important reference indicators for assessing the degree of vascular aging. Moreover, this method may benefit the research on age-related cardiovascular diseases. Nevertheless, it should be noted that the SHG intensity was highly dependent on the overlap of the laser polarization with the fiber alignment. The excitation light used in this study was linearly polarized. The intensity of the SHG signal appeared to be at a maximum when the laser polarization direction was parallel to the orientation of the collagen fibers, whereas it appeared at a minimum when the two directions were perpendicular. We hope to consider laser polarization in our future studies, despite a variety of measures taken to minimize the effects of polarization on the quantitative analysis results of the SHG images in the present study. In addition, we found that the three age groups considered in this study could not be completely distinguished from each other by relying merely on a single 3D GLCM texture feature, although the 3D GLCM algorithm is considered highly sensitive to fiber microstructures. Therefore, more sensitive and valuable quantitative analytical methods merit further investigation.
SignificanceOral cancer is among the most common cancers of the head and neck. Despite advancements in targeted cancer therapy, the survival rates of oral cancer patients have plateaued over the last 50 years. Common screening methods for oral lesions, such as visual inspection and palpation of tissue surfaces, are highly dependent on the experience of clinicians. Even if the biopsy or histopathological examination is performed for highly suspicious tissue regions, the limitations of time-consuming, invasive, and label-intensive are still inevitable. In clinical practice, intraoperative frozen section biopsies for surgical margins are routine procedures performed after en bloc resections of oral cancers. However, surgical margins are usually selected according to surgeon estimates of sites that may be suspicious of inadequate resection, resulting in the omission of positive margins. In addition, early detection of oral cancer plays a critical role in improving the prognosis and survival rate, but, accurate identification is difficult based on conventional screening methods.To improve the clinical diagnosis of oral diseases, researchers have conducted numerous studies on auxiliary diagnostic techniques, including X-ray computed tomography (X-CT), magnetic resonance imaging (MRI), ultrasound imaging (UI), fluorescence imaging (FI), photoacoustic imaging (PAI), and optical coherence tomography (OCT). Based on the associated imaging theories, different imaging technologies have unique advantages in terms of detecting oral diseases, resulting in different application scenarios. In this paper, we review the research on the foregoing auxiliary imaging technologies, summarize their advantages and disadvantages, and discuss the challenges and future developments in oral clinical applications.ProgressDifferent technologies demonstrate different features in terms of improving diagnostic sensitivity, specificity, resolution, and so on. Notably, X-CT and MRI are the earliest techniques used in oral clinics. They are exceptional in terms of their imaging depth and can evaluate bone invasion and the thicknesses of oral cancers.In recent years, with improvements in ultrasonic technology, the imaging resolution of UI using ultra-high-frequency ultrasound (30-100 MHz) has considerably improved. Such improved resolutions facilitate the observations of smaller microstructures (approximately 30 μm in size) of oral tissues. One recent study demonstrated that diagnostic sensitivity, specificity, and negative predictivity with values of over 90% were achieved in 150 patients with oral soft tissue lesions using an ultra-high UI system. In addition, Doppler ultrasound plays a major role in evaluating the neovascularization of oral neoplasms and metastatic lymph nodes by obtaining blood flow information (Fig. 3).Advancements in FI, including both auto- and extrinsic fluorescence, have enabled the exploitation of molecular information. Interestingly, autofluorescence of the oral epithelium and submucosa can be generated by laser excitation at 400-460 nm, which can then be used to identify oral lesions derived from changes in the concentration and properties of fluorophores. In contrast to benign oral mucosal lesions, malignant lesions are associated with autofluorescence loss. However, several benign lesions also exhibit fluorescence decay, resulting in low specificity. Through the continual exploration of fluorescent dyes and targeted tumor biomarkers, FI can achieve higher specificity in the detection of oral tumors.PAI is an imaging technology that has undergone developments in recent years and is based on the photoacoustic effect. Combining the advantages of optics and ultrasound, this technique has technical advantages in detecting oral tumor neovascularization (Fig. 5).OCT, which is a high-resolution, non-destruction, and label-free method, has been successfully used in ophthalmology, cardiology, and gastroenterology. Moreover, the feasibility of OCT in distinguishing different oral tumors has been verified (Fig. 6). In addition, for the early detection of oral cancer, OCT has been used to detect different types of oral mucosal leukoplakia (Fig. 7).To facilitate oral clinical studies, PAI and OCT are also undergoing rapid developments in terms of system miniaturization. In recent years, researchers have developed various miniaturized probes for oral imaging (Fig. 9).To compensate for the shortcomings of single-imaging techniques, multi-modal systems combining multiple diagnostic techniques have also been developed.With visual observations or qualitative analysis, misdiagnosis is inevitable. To improve the accuracy of image recognition and reduce the time cost associated with image reading, quantitative analysis and artificial intelligence approaches based on oral tissue images have been widely studied with the aim of extracting rich information from images (Fig. 10).Conclusions and ProspectsImaging technologies with non-destruction, high resolution, high sensitivity, high specificity, and real-time will play a critical role in assisting clinicians in screening and diagnosing oral cancers. Owing to the unique characteristics of different imaging techniques, their clinical application scenarios are different. Single-imaging techniques cannot completely satisfy all the requirements of oral disease diagnoses. Therefore, combining multiple imaging techniques to construct a multi-modal system can provide more abundant diagnostic information. In addition, quantitative and AI-based computer-aided methods that can provide objective screening and diagnostic results are expected to be developed.
ObjectiveThe increasing burn mortality rate places an urgent need for accurate diagnosis and treatment of burns. Currently, the third-degree quartile is internationally used to classify the degree of burns based on burn depth, and clinical treatment methods for different degrees of burns are significantly dissimilar. Burn surgeons overestimating the severity of burns can lead to unnecessary surgery, whereas underestimating them leads to treatment delays and worsening of the burn conditions. In addition, studies have shown that burn severity changes dynamically over time, with superficial Ⅱ burn worsening to deep Ⅱ or Ⅲ burns within 48 h of burn occurrence. Therefore, overcoming the defects of subjective judgment using the naked eye and quantitatively monitoring the dynamic changes in the burn degree in real time has become a challenge in the early diagnosis of burns. Burn diagnosis methods based on photonics, such as near-infrared spectroscopy, reflective confocal microscopy, and laser Doppler flowmetry, are developing rapidly. However, their clinical application is limited owing to low accuracy, invasiveness, high detection environment requirements, and high costs. In this study, a noninvasive quantitative method for assessing the burn degree was developed based on spatial frequency-domain imaging (SFDI). Combined with the systematic clustering method and multiparameter dimensionality reduction analysis, the proposed method results in improved classification accuracy of different burn degrees and shortened classification time, thus indicating the potential for early diagnosis of clinical burns.MethodsIn this study, the SFDI technique was applied to a rat burn model. First, the backs of Sprague-Dawley (SD) rats were depilated, and a thermostatic iron heated to 100 ℃ was used on the backs of the anesthetized SD rats for 4, 12, and 24 s, respectively, to establish a rat burn model with different burn degrees. Next, the sinusoidally modulated structural patterns were projected onto the surface of each burned area, and the backscattered structural patterns from the tissues were captured using a charge-coupled device (CCD) camera. Subsequently, we used single-snapshot multifrequency demodulation (SSMD) to extract the modulation transfer function (MTF) of light from the burned tissues. Compared with the traditional three-phase shift demodulation method, SSMD only requires a single snapshot to achieve parameter extraction, which significantly suppresses the problem of motion artifacts and improves the signal-to-noise ratio of imaging using filtering technology. Based on the photon diffusion transmission theory, the optical parameters (μa and μ′s) were then recovered using the look-up table method at the 5th, 10th, 30th, 60th, 90th, and 120th minutes after burn. Finally, systematic clustering and multiparameter dimensionality reduction analysis were performed on the optical parameters to quantify and classify different burn degrees.Results and DiscussionsDifferent degrees of burns can be effectively distinguished by the relative changes in the two optical parameters at the three wavelengths. The results show that the magnitude of the absorption coefficient positively correlates with the degree of burn. In contrast, the magnitude of the reduced scattering coefficient negatively correlates with the degree of burn. Although the distinction between optical parameters is not significant at the beginning of burns, the optical parameters of the 4 s burn group gradually decrease or gradually recover to the unburned state with observation time. In contrast, the optical parameters of the 12 s and 24 s groups gradually deviate from the normal state (Fig. 6). The burn results are divided into two categories through optimal analysis of systematic clustering. The 4 s group is classified as mild burns, whereas the 12 s and 24 s groups are classified as severe burns. Although the classification accuracy is less than 85% in the first 10 min after burn, it is 100% in the later stages (Table 1). Two new factors (the absorption factor FAC1 and the reduced scattering factor FAC2) reflecting approximately 93% of the original variable information can be generated using the principal component analysis to reduce the dimensionality of the six optical parameters. The results show that the absorption factor, FAC1, distinguishes the degree of burns in a large category (mild burns in the 4 s group and severe burns in the other two groups) and increases the difference between deep Ⅱ degree burn in the 12 s group and Ⅲ burn in the 24 s group. In addition, the assessment of burn severity using principal constituent factors can reduce interference and improve classification accuracy in the early stage after burn (Fig. 9).ConclusionsThe quantitative burn imaging device based on real-time spatial frequency-domain imaging technology has remarkable advantages over existing diagnostic techniques, for example, ease of handling, compact structure, and high precision. Through dynamic monitoring of changes in optical parameters combined with cluster analysis and parameter dimensionality reduction, the degree of burns can be determined through noninvasive assessment, providing a reliable guarantee for the precise treatment of burns. In future studies, we will supplement the pathological verification, characterize additional physiological parameters (such as hemoglobin content, blood oxygen saturation, and melanin concentration) from the optical parameters, and extend this technology to clinical applications so as to significantly reduce the treatment cycle and cost to patients.
ObjectiveFluorescence molecular imaging is widely used in clinical practice. Different hardware and software designs in different fluorescence imaging systems lead to differences in imaging performance between instruments. However, unlike radiographic imaging, fluorescence imaging currently has no mature specifications or standards to test the performance of imaging instruments. Phantoms are tools used in standardized imaging performance testing and are commonly used in radiography. Unlike human tissues, phantoms have preset shapes and contrasts that simulate specific tissue parameters over time. These properties allow phantoms to be used to measure, evaluate, and confirm the performance of imaging instruments. In research on near-infrared fluorescent phantoms, fluorescent agents and quantum dots are used as samples for fluorescence imaging performance testing. However, owing to the different materials and preparation methods used in the samples, as well as the stability of the materials themselves, there is still no stable sample for accurately simulating the fluorescence spectrum as a standardized test tool. In this study, a fluorescence emission simulation system for simulating fluorescent samples is proposed. Imaging instruments can exhibit the same response as real fluorophores when imaging a fluorescence simulation system. Compared with traditional fluorescent agents, the fluorescence emission simulation (FES) system can more accurately simulate fluorescence emission characteristics in a stable manner.MethodsIn this study, we propose a fluorescent light-emitting system that uses an optical system to simulate fluorophores. First, according to the characteristics of the fluorescent agent, a simulation method for fluorescence excitation efficiency, spatial distribution, and fluorescence emission spectrum characteristics is designed such that the fluorescence imaging instrument has the same response as the real fluorescent agent when imaging the FES system. The system controls the intensity of the outgoing fluorescence according to the intensity of the detected excitation light, thereby simulating the fluorescence excitation efficiency of the fluorescent agent sample. The design of the optical entrance and exit based on the integrating sphere can simulate the spatial distribution of outgoing fluorescence. In this study, a spectral simulation method based on a linear filter LVF and liquid crystal display (LCD) is used, and based on this method, an improved least squares spectral fitting algorithm is designed to automatically simulate arbitrary fluorescence spectra.Results and DiscussionsThe performance verification shows that the subspectra of the FEM system have linear additivity (Fig. 5), and the subspectrum satisfies a certain gray-transmittance relationship [Fig. 6(a)]. A grayscale transmittance curve of the sub-spectrum was obtained [Fig. 6(b)]. Functional verification of the system is realized by fluorescence emission simulation of ICG. The simulation of the fluorescence emission spectrum characteristics [Fig. 7(a)] and fluorescence excitation efficiency [Figs. 7(b)-(d)] of the ICG aqueous solution samples with different concentrations is performed using the FES system. The simulation system obtains the same test results as the fluorescent samples (Fig. 8), and compared with traditional fluorescent agents, the FES system more accurately simulates the fluorescence emission characteristics in a stable manner, which verifies the feasibility of the system as a standardized test tool.ConclusionsIn this study, a method for simulating fluorescent samples with an optical system is proposed. A programmable FES system is built and an optical system is used to simulate fluorescent agents for the standardization test of near-infrared fluorescence imaging performance. The FES system can simulate the fluorescence excitation efficiency, spatial distribution, and fluorescence emission spectrum characteristics of the fluorescent sample such that the fluorescence imaging instrument has the same response as the real fluorescent agent when imaging the FES system. A spectral simulation method based on a linear filter and liquid crystal display is also proposed and an improved least squares spectral fitting algorithm is designed, which can automatically simulate any fluorescence spectrum. Finally, based on the FES system, the fluorescence imaging sensitivities of different near-infrared fluorescence imaging instruments are tested and the test results of different instruments are compared. The test results show that compared with traditional fluorescent agents, the FES system can more accurately simulate the fluorescence emission characteristics in a stable manner, which verifies the feasibility of the system as a standardized test tool.
ObjectiveQuantitative measurement of burn depth is of great significance for the clinical assessment of burn degree and treatment plan. Currently, the most widely used assessment method is visual inspection, which places high demands on doctors’ experience and is easily influenced by subjective judgment. Other detection techniques, such as laser Doppler imaging, ultrasound imaging, and fluorescence imaging, have also been used to assess the extent of burns; however, these techniques cannot non-invasively and accurately measure burn depth. Polarization-sensitive optical coherence tomography (PSOCT) has the advantages of non-invasiveness, fast imaging speed and high resolution and can quantitatively measure the burn depth based on the polarization information of the burned tissue. However, the traditional measurement method is based on the accumulated polarization information from the sample surface to a certain depth inside the sample, which cannot accurately characterize the local polarization information at this depth; hence, the burn depth cannot be accurately measured. Therefore, this study proposes a local polarization information extraction algorithm based on spectral domain polarization-sensitive optical coherence tomography (SD-PSOCT) to obtain polarization information at each depth inside the burned biological tissue to quantitatively measure the burn depth of the biological tissue.MethodsA local polarization property extraction algorithm based on the SD-PSOCT system was proposed and used to quantitatively measure the burn depth of biological tissue. All single-mode-fiber-based systems adopt fiber-based polarization controllers to illuminate a sample with a single-input polarization state. A custom-built linear-in-wavenumber spectrometer consisting of a diffraction grating, dispersive prism, Wollaston prism, and a focusing lens was used to realize polarization-sensitive detection (Fig. 2). Then, the local phase retardation and axis orientation of each layer of the sample were calculated by eigenvalue decomposition based on the Jones matrix and layer-by-layer iterative algorithm. To evaluate the measurement accuracy and stability of the system, we used a quarter-wave plate (QWP) as the sample and measured the phase retardation and axis orientation of the QWP under different axis orientations each day for 14 days. To measure the burn depth of the biological tissue, we selected a piece of bovine tendon tissue as experimental sample, burned the same position of the bovine tendon five times for 10 s each, and then reconstructed the local phase retardation images of the bovine tendon unburned and burned for 10 s, 20 s, 30 s, 40 s, and 50 s, respectively. We then considered the full width at half maximum of the local phase retardation versus the imaging depth curve as the burn depth.Results and DiscussionsFrom the sensitivity roll-off curves we can see that the sensitivity at the detection depths of 0.2 mm and 1.2 mm are approximately 105 dB and 98 dB, respectively (Fig. 3). The measured average value of the phase retardation of the QWP is 82.9° and the measurement error is 1.9° (Fig. 4). The 14-day measurement results show that the phase retardation varies within a range of -0.42° to + 0.42° and the axis orientation varies within a range of -0.66° to + 0.66°. By comparing the local phase retardation images of the bovine tendon subjected to different burn times (Fig. 6), it is found that the local phase retardation inside the burned bovine tendon increases, and as the burn time increases, the region with a higher local phase retardation extendes to a deeper position. From the depth-resolved local phase retardation, it can be seen that the region with higher phase retardation gradually widens with increasing burn time (Fig. 6). Thus, the measured burn depth of bovine tendon tissue burned for 50 s is 390 μm.ConclusionsWe deduce the local polarization property extraction algorithm based on the Jones matrix in detail and provide the calculation formulas of local phase retardation and axis orientation. The sensitivity roll-off curves of the two orthogonal polarization channels in the linear wavenumber spectrometer are experimentally measured, and the measured sensitivity of the system is 105 dB. The actual phase retardation and axis orientation of the QWP at different axis orientations are measured and it is verified that the system can measure the polarization properties of birefringent samples with high accuracy and maintain good measurement stability. The imaging results of bovine tendon tissue subjected to different burn times show that the SD-PSOCT system can obtain polarization images with higher contrast than traditional OCT images. Additionally, compared with the cumulative phase retardation image, the local phase retardation image obtained by the algorithm can highlight the difference in the bovine tendon after being burned for different times and quantitatively measure the burn depth according to the local phase retardation images. This study provides a new method for quantitatively measuring tissue burn depth, which can be applied to clinical diagnosis and burn treatment in the future.
ObjectiveDetection of phytoplankton diversity is an important part of a water quality bioassessment. The traditional manual microscopic detection of algae species requires professional operation and is time-consuming and laborious; therefore, these challenges can be overcome by the development of a method for automatic identification of phytoplankton cell images. Similar to manual identification, deep learning and other automatic identification technologies identify phytoplankton cells based on the morphological characteristics of bright field cell images; however, the practical applications of such technologies encounter complications, such as the difficulty of accurate segmentation of phytoplankton cells and the limitation in high recognition accuracy of algae species only with a small range of groups. Previous studies have shown that the accuracy of algal cell segmentation and recognition can be effectively improved by fusing the bright field and fluorescence images of phytoplankton cells. However, the fusion of bright field and fluorescence images synchronously measured from phytoplankton cells requires significantly high accuracy and shockproof capability of the mechanical structure of the acquisition system. Under a high-power microscope, even a small error in the mechanical structure or a small vibration of the camera can lead to a dislocation between the bright field and fluorescence images, thus causing difficulties in the fusion of the images. Therefore, the registration of bright field and fluorescence images of algae is of great significance for the automatic identification of phytoplankton.MethodsThe displacement between bright field and fluorescence images can be represented by a rigid transformation model, which consists of three parameters: the translation in the x direction, translation in the y direction, and rotation angle. Normalized mutual information is used to calculate the similarity between the bright field and fluorescence images. The goal of image registration is to find a set of rigid transformation parameters that maximize the normalized mutual information of the two images. Owing to the significant difference between the bright field and fluorescence images of phytoplankton cells, the similarity is difficult to be characterized by directly calculating the normalized mutual information. In this study, the normalized mutual information of the S channel binary image of the bright field HSV color space and the binary image of the fluorescence gray level was assumed as the similarity between the bright field and fluorescence images. The S component of the bright field HSV image and the fluorescence gray image were decomposed using a two-dimensional discrete wavelet transform, and the low-frequency components were binarized, to accelerate the registration. First, the particle swarm algorithm was used to register the low-frequency components of the five-level wavelet decomposition. Subsequently, the translation and rotation angle of the preliminary registration were assumed as the initial values, and the low-frequency components of the wavelet three-level decomposition are further registered using Powell’s algorithm.Results and DiscussionsScenedesmus sp., Selenastrum capricornutum, and Nostoc sp. are used as experimental objects. The similarity and registration methods are compared and analyzed. As shown in Figs. 3 and 4, the normalized mutual information of the bright field S channel and the fluorescence grayscale image has an obvious peak value after binarization, which is extremely conducive to parameter optimization in the registration process. As shown in Fig. 5, after the bright field and fluorescence images are decomposed by the wavelet, the noise in the high-frequency component is concentrated, and the low-frequency component has a higher normalized mutual information. Therefore, the normalized mutual information of binary images of low-frequency components after wavelet decomposition is chosen as the similarity measurement index for bright field and fluorescence images. Table 1 presents an experimental comparison between the proposed method and other registration methods. Compared with particle swarm optimization algorithm, the proposed method reduces the mismatch rate by 0, 8.1, and 0.7 percentage points, shortens the running time by 128.26 s, 448.95 s and 237.20 s, and improves the normalized mutual information by 0.065, 0.083, and 0.106; Powell’s method depends on the initial value; therefore, it is easy to fall into the local maximum in the process of optimization, which leads to a high mismatch rate; compared with GA, the mismatch rates of proposed method are reduced by 6.1, 26.2, and 10.2 percentage points, the running time is shortened by 23.78 s, 60.95 s and 33.74 s, and the normalized mutual information after registration is improved by 0.149, 0.170, and 0.180. The experimental results demonstrate that the proposed method has obvious advantages in terms of registration accuracy and running time compared with other registration methods.ConclusionsIn this study, the bright field image is converted into a binary image by processing the S channel of the HSV color space; the fluorescence gray image is binarized; normalized mutual information is used as the similarity criterion to characterize the similarity of the bright field and fluorescence image. Using Scenedesmus sp., Selenastrum capricornutum, and Nostoc sp. as experimental objects, the application of wavelet decomposition in the registration of microscopic bright field and fluorescence images of phytoplankton cells was studied. The global search advantage of the particle swarm optimization algorithm is used for preliminary registration in the high-level decomposition component, and the local search ability of Powell algorithm is used for fine-tuning the registration accuracy in the low-level decomposition component. A comparative analysis is performed with other commonly used registration methods, and the experimental results verify the feasibility of the registration method.
ObjectiveDiabetes is one of the major diseases affecting human health. Patients are required to monitor and control their blood glucose concentration continuously to prevent complications. Compared with traditional invasive methods, optical noninvasive blood glucose detection greatly reduces the pain of blood collection for patients and controls blood glucose more accurately by increasing the number of measurements. Recently, the noninvasive blood glucose detection method using optical coherence tomography (OCT) has become a technology with great development potential. However, the acquired three-dimensional images in skin areas are inconsistent in spatial range during the image acquisition due to the slight involuntary shaking of the subject, affecting the accuracy of the noninvasive blood glucose measurement. To solve this problem, we propose a skin three-dimensional image correction and compensation method based on OCT noninvasive blood glucose detection.MethodsThe deviation of the 3D OCT image collected during noninvasive blood glucose detection is corrected and compensated using an algorithm. First, we study the influence of image deviation on experimental data using theoretical analysis. In the experiment, the skin area is roughly positioned by pasting a square positioning label on the skin surface. Due to the uneven border of the positioning and the involuntary movement of the arm when collecting images, a deviation is observed in the image of the extracted skin area, leading to errors in the calibration and prediction results. The image of the skin area is extracted using morphological erosion expansion, then the image boundary is smoothed and the skin surface is aligned. After the template is determined by selecting the central area of the collected skin image, the compensation algorithm is used to gradually match the template with the skin area images involved in the subsequent blood glucose calculation and perform correction and compensation to ensure consistency in the skin area. We compare the data before and after compensation by the human blood glucose experiments. Furthermore, we compare the predicted results before and after compensation, such as the standard deviation of the actual and predicted blood glucose concentration, correlation coefficient, and other parameters, to validate the compensation algorithm.Results and DiscussionsIn this paper, the deviation of the skin image data is corrected using a compensation algorithm. Figure 6 shows a comparison between the skin images acquired at different moments during the blood glucose collection and the image corrected using the template image. The results show a high correlation between the template and correction images. The average correlation coefficient between the image and template is 7.3 times higher than that without correction. The number of relevant areas in the calibration chart after compensation increases, as shown in Table 2. Furthermore, we obtain the difference between the prediction results before and after the compensation. From the prediction chart, the prediction results calculated from skin image after applying the compensation algorithm are closer to the real blood glucose concentrations. Table 3 shows the correction and compensation effects of different volunteers. The correlation coefficient increases from 0.37 to 0.65, and the proportion of predicted blood glucose in zone A using the Clark grid error analysis increases from 51.82% to 79.86%. Additionally, the standard deviation of the prediction results decreases from 1.34 mmol/L to 0.83 mmol/L after compensation. Clinical experimental data show that the accuracy of the noninvasive blood glucose prediction value is improved by 36.57%. Moreover, we study the influence of the change in the blood glucose concentration on the scattering coefficient. The results show that the scattering coefficient change caused by the blood glucose concentration change of 1 mmol/L is about 15.8 times greater than that when there is no blood glucose change.ConclusionsAs a methodology of optical noninvasive blood glucose detection, OCT is noninvasive, real-time, and sensitive to changes in blood glucose concentration in dermal tissue interstitial fluid. In this paper, we propose a compensation algorithm for image correction to improve the motion artifacts caused by the subject shaking. The compensation algorithm can find the position with the greatest similarity between the template and each image. The results show that using the compensated skin image for blood glucose calibration calculation increases the number of areas with higher correlation coefficients in the calibration chart. Meanwhile, the deviation of the predicted result is smaller than that without compensation. Furthermore, the difference between the predicted and actual values decreases after compensation, and the correlation is better than that before compensation. Therefore, the compensation algorithm effectively solves the prediction error caused by the extracted skin area deviating. This study has the important reference value for optical noninvasive blood glucose detection and is suitable for other optical imaging applications that require accurate quantitative calculations.
ObjectiveBreast cancer has been the most common life-threatening cancer of women in recent years. Both diagnostic imaging and pathology are routinely employed to diagnose breast cancer, while the latter is considered the "gold standard" for cancer diagnosis. In clinical practice, however, the routine pathological diagnosis is strongly hindered by the complicated and time-consuming process of staining the biopsy sections with hematoxylin and eosin (H&E) for highlighting the fine structures of cells and tissues. Moreover, the diagnosis is manually performed by pathologists, therefore the diagnostic speed and accuracy are highly dependent on their knowledge and experience. In order to better serve the treatment of breast cancer, the pathological diagnosis is greatly expected to be accelerated and automated, especially providing the efficient intraoperative assessment for precision surgical therapy. In this study, a rapid, accurate and automatic diagnostic technique of breast cancer is proposed based on the polarization and bright-field multimodal microscopic imaging. To accelerate the pathological diagnosis, the breast biopsy sections are directly investigated without the H&E staining, while the cell structures are no longer distinct under the bright-field microscopic imaging. In this case, the polarization microscopic imaging is introduced to further extract the morphological difference between the normal and cancerous breast tissues. Collagen fiber is an important part of the extracellular matrix (ECM), and the organization of collagen fibers has been found to be closely related with the cancer progression. Due to the inherent optical anisotropy, collagen fibers can be examined by cross-polarization imaging. To perform the automatic diagnosis, deep learning is employed to distinguish the normal and cancerous breast tissues, where the convolutional neural network (CNN) classification model is established to extract features from the multimodal microscopic images and make the accurate and reliable judgements.MethodsIn this study, 23 breast biopsies from 16 patients were first rapidly frozen in liquid nitrogen and cut into sections with the thickness of 15 μm using a cryotome. Without the H&E staining, the bright-field microscopic imaging and polarization microscopic imaging were sequentially performed through switching the light source, polarizer and analyzer of the custom-made transmission polarization microscope. The polarization microscopic imaging was operated in the cross-polarization mode, where the polarizer was orthogonal to the analyzer. Since the period of cross-polarization imaging is 90°, the polarizer-analyzer pair was rotated by 0°, 30° and 60° to characterize the biopsy sections. Following the microscopic imaging, the pixel-level image fusion was conducted to transform the four multimodal images into a single fused image for breast cancer diagnosis. In order to perform the deep learning-assisted automatic diagnosis, the classical CNN ResNet34 was utilized to develop a classification model, whose input is the pixel-level fused image. In addition to the pixel-level fusion, the decision-level fusion was also examined for comparison, where two classification models were created based on the bright-field and polarization microscopic imaging, respectively. In this way, the final classification result was generated according to the weight coefficients determined using the logistic regression algorithm. To evaluate the performance of CNN classification model, four parameters including accuracy, sensitivity, specificity and AUC [area under receiver operating characteristic (ROC) curve] were calculated. Due to the rather small amount of breast biopsies, the leave-one-out cross validation was performed in order to avoid overfitting.Results and DiscussionsThe cross-polarization microscopic imaging of normal breast tissues exhibits the distinct periodic change in the brightness with the rotation of the polarizer-analyzer pair from 0° to 90°, while that of cancerous breast tissues is almost unchanged (Fig. 3). This polarization-sensitive brightness change results from the anisotropic organization of collagen fibers in the normal breast tissues, which alters during the cancer progression. Further, the classification result of the multi-polarization imaging is better than that of the single-polarization imaging (Table 1). Following the pixel-level fusion of multimodal images, the CNN classification model based on ResNet34 was established. As a result, the accuracy of 0.8727 and AUC of 0.9400 were realized, better than bright-field (0.8540, 0.9013) and polarization (0.8575, 0.9307) microscopic imaging, respectively (Table 2). In addition, the decision-level fusion was also evaluated, achieving the accuracy of 0.8710 and AUC of 0.9367. The weight coefficients of bright-field and polarization imaging were calculated to be 0.3971 and 0.6029, respectively.ConclusionsA deep learning-assisted multimodal microscopic imaging technique is proposed for the rapid, accurate and automatic diagnosis of breast cancer, combining bright-field and cross-polarization microscopic imaging. In this scheme, the time-consuming process of H&E staining can be removed to accelerate the breast cancer diagnosis, and the automatic diagnosis can be performed with the CNN classification model. Moreover, the accurate diagnosis can be realized, since the cross-polarization microscopic imaging can further extract the polarization-sensitive morphological change closely related with the cancer progression, such as the organization of the optically anisotropic collagen fibers. In this sense, the multimodal microscopic imaging diagnosis has great potential to better serve the surgical treatment of breast cancer with the efficient intraoperative assessment.
ObjectiveIn the process of medical image acquisition, due to some factors of the image acquisition device (such as improper parameter adjustment and the limitation of the equipment’s inherent attributes) or the conditions of the object itself (that is, the light absorption and reflection of different attributes) makes the signal collecting process and transferring process in the presence of complicated noise model, causing lung CT image has the characteristics of low contrast and visible mask. Therefore, images with poor visual quality seriously interfere with the efficiency of clinical diagnosis and are a significant obstacle to the subsequent use of images. There is a lot of research on medical image enhancement, but the work on lung CT image enhancement is still lacking. Additionally, when processing images, existing image contrast enhancement algorithms based on histogram equalization tend to introduce unnecessary artifacts, produce an artificial appearance, and cause wash-off effects. Therefore, this paper researched lung CT image enhancement.MethodsWe devote to overcoming this over-enhancing problem of existing algorithms and then propose an algorithm which can realize appropriate contrast enhancement without introducing new artifacts, that is an image enhancement algorithm based on image segmentation and a total variation model. As is known to all, lung CT images are poor in contrast due to their narrow dynamic grayscale range. And the visual perception of difference relies on gray histogram distribution characteristics to a great extent. Therefore, the research method of contrast enhancement adopted in this article is based on gray histogram transformation. Furthermore, regarding the feature differences between the foreground and background of lung CT images, a segmentation method based on a global threshold is used to segment the lung parenchyma that doctors are interested in for further processing.As for the complex noise model in the image, traditional denoising methods are challenging to ensure the regularization of image enhancement results. Consequently, this framework uses the gradient descent method and the total variation framework to separate the image’s noise from the perspective of minimizing energy. Following that, the image structure information and the image detail information will be obtained along with the noise. Then, wavelet transform technique is used to suppress the noise among the image detail information.The pipeline of the algorithm is as follows, dividing the image into foreground and background firstly, performing bipolar threshold clipping and cumulative distribution function redistribution on the histogram of the foreground lung parenchyma image to form a modified histogram, and then performing Gamma adaptive stretching on the image according to the modified histogram. As a result, the contrast-enhanced foreground image is obtained and fused with the background image as the input of the total variation model. The total variation model then decomposes the image into a texture layer and a structural layer. Next, the texture layer is denoised by the wavelet threshold. Finally, the enhanced image is obtained by fusion of texture layer and structural layer.Results and DiscussionsThis research proposed a framework for improving lung CT images using image segmentation, histogram modification, total variational, and wavelet transform technique (Fig. 2). The subjective analysis of the experimental results shows that the algorithm effectively suppresses the artifacts noise of the image, solves the defect of the existing algorithm over-enhancing lung CT image, comprehensively improves the image contrast, and preserves the complete natural information of the image, as shown in Fig. 3 and Fig. 4. The comparison of image details in Fig. 3 shows that the enhanced image maintains a reasonable degree of regularity in terms of appearance display, texture details, and edge characteristics.The average value of the objective evaluation index of the experimental results is shown in Table 1. It can be seen that the objective evaluation index parameters of the proposed method have obvious advantages compared with other existing image enhancement methods by comparing the image evaluation index, such as the contrast, grayscale resolution, structural similarity, and absolute mean brightness difference. For instance, the proposed algorithm framework not only fully enhances contrast by increasing grayscale dynamic range display, but also assures the regularity of the enhanced results. The average intensity of the enhanced image by our algorithm is closest to that of the original image, showing the enhanced image has the highest similarity with the original image.ConclusionsThis paper proposes an image enhancement algorithm that solves the issues of low contrast and visible mask in lung CT images. Furthermore, it overcomes the tissues of over enhancement and washout effects which are easy to occur in existing image enhancement algorithms. The research shows that the proposed method can effectively suppress the artifacts noise of the original image in the different areas of the test image, enhance the contrast of the lung CT image, improve the visual effect significantly, and overcome the unwanted artificial artifact greatly. The algorithm is significantly better than other enhancement algorithms in terms of subjective performance evaluation and objective evaluation index. Therefore, the enhancement framework proposed in this paper can provide robust technical support for lung CT image enhancement and improve the efficiency and accuracy of clinical diagnosis and treatment.
ObjectiveCancer is a disease caused by the uncontrolled growth and division of malignant cancer cells. Since the 21st century, the incidence and mortality of cancer have been increasing rapidly worldwide, making it a medical problem that affects countries worldwide. The pathological mechanism of cancer and the study of various therapeutics based on inducing cancer cell death both greatly benefit from research on the morphology and function of cancer cells at the single-cell level, particularly research on the process of cancer cell death. Digital holographic microscopy, a quantitative phase imaging technique, offers a nondestructive, unlabeled, and noncontact quantitative measurement tool for biological research. It can also provide nondestructive quantitative imaging of living cells. In this paper, digital holographic tomography was used in the three-dimensional quantitative detection of bladder cancer cell vacuolation. This work can broaden the field of application for digital holographic tomography in the biomedical industry, offer new perspectives on how to study the morphological changes that occur during cancer cell apoptosis, and investigate potential new cancer treatment approaches.MethodsThis study focuses on the vacuolar structure of cancer cells existing in the process of paraptosis. First, the hologram of bladder cancer cells with vacuoles inside was obtained using digital holographic microscopy. Then, the amplitude and phase of cells were obtained by filtering, digital focusing, angular spectrum propagation, and phase unwrapping. Their three-dimensional morphology and spatial locations were reconstructed using the diffraction tomography reconstruction algorithm combined with nonnegative constraints. Finally, morphological parameters such as the surface and volume of the vacuoles were calculated according to the number of pixels in the image.Results and DiscussionsDigital holographic tomography was used in this study to produce three-dimensional reconstruction results of four bladder cancer cells with vacuoles (Figure 8). Four morphological parameters, including the volume, surface, surface to volume, and the ratio of vacuoles volume to cell volume, were calculated (Table 1). Digital holographic tomography, as a technique for three-dimensional quantitative imaging, was used to examine cancer cells with vacuoles. It can quantitatively determine the volume, position, and other morphological parameters of the vacuoles. By combining with the biomedical research, it can be used to observe the changes in three-dimensional shape and volume of tumor cells’ internal vacuoles induced by drugs, to explore the correlation between the expression of some proteins and the morphological characteristics of vacuoles, to provide a more comprehensive and profound understanding of the process of paraptosis of cancer cells and to find new methods for cancer treatment.ConclusionsThe application of digital holographic tomography to bladder cancer cell vacuolation imaging is described in this research. The results have shown that digital holographic tomography can accurately reconstruct the three-dimensional shape and space position of the vacuolation in bladder cancer cells. The above research progress is of great significance for studying the paraptosis process of cancer cells as well as the related mechanisms and treatment strategies of drug-induced paraptosis of cancer cells.
SignificanceMedical imaging is a key tool for life science research, diagnosis and treatment. Traditional medical imaging techniques include magnetic resonance imaging (MRI), computed tomography (CT), positron emission tomography (PET), ultrasonic imaging (US) and optical coherence tomography (OCT), etc. Each of them plays an important role in medical imaging, with each showing its own advantages and limitations. MRI is not only expensive but also has many safety restrictions, which means it is not suitable for the patients with pacemakers or claustrophobic. CT is not suitable for children and pregnant women because of ionizing radiation. PET has a large number of applications in the study of life metabolism and tumor research. However, it requires radioactive element markers and is difficult to be widely used in clinical medical imaging due to its high cost. US has poor specificity and spatial resolution, and lacks fine biological structure imaging capabilities for the early development of the disease. OCT and other optical imaging technologies are limited by the optical diffusion limit, which results in the general imaging depth within about 1 mm.Photoacoustic (PA) imaging is a hybrid biomedical imaging modality combining the advantages of high contrast of optical imaging and deep penetration of ultrasound imaging. The spatial scale of PA imaging covers from subcellular structures to organs. In addition, it has many other advantages such as non-invasive imaging, label-free imaging, molecular imaging and compatible with multi-modality. Although PA imaging has encountered many challenges in the process of clinical translation, PA imaging has overcome a series of difficulties and will have broader application prospects in the field of biomedical imaging thanks to the development of related technologies. The purpose of this article is to help readers in related fields of biomedical imaging to form a more comprehensive understanding of PA imaging, and to quickly understand the main progress of PA imaging research in recent years.ProgressThis review article provides a brief introduction to the basic principles and main modes of PA imaging. Photoacoustic computed tomography (PACT) and photoacoustic microscopy imaging (PAM) are the two main modes of PA imaging. Photoacoustic endscopy (PAE) is the application of PA in endoscopy, and photoacoustic molecular imaging expands the capabilities of PA imaging in molecular-level detection.PACT is suitable for large-scale imaging of the human brain, limbs, breast and other large-size targets. Over the past decade, PACT has made lots of advances in high-speed and deep imaging. However, the issues about economy and portability still hinder the further clinical translation of PACT. Fortunately, the technological development of ultrasonic transducers and low-cost laser sources and the advancement of advanced reconstruction algorithms have provided solutions to the above problems. PAM has broad application prospects from subcellular structure to organ level. High/super-resolution, fast imaging, and higher imaging quality have always been the common pursuit of researchers. The nonlinear effect greatly contributes to the improvement of the resolution of PA imaging. The extended depth-of-field technology can solve the defocus problem encountered by PAM in volume imaging. Advanced scanning methods are one of the main solutions to high-speed PA imaging. In addition, non-contact PA imaging is another important direction in clinical applications, and corresponding technological breakthroughs have also been made in recent years. As the expansion of PA imaging, PAE is a promising technology for endoscopic imaging by drawing on the related progress of PACT and PAM, such as extended depth-of-field technology, optical scanning methods and multi-modality imaging. Aiming at the problem that some tumors lack characteristic absorption peaks, PA molecular imaging holds great promise in the diagnosis and treatment of diseases. In recent years, the development of PA molecular imaging has focused on the near-infrared window to improve the depth of imaging. The PA signal enhancement mechanism can improve the sensitivity and specificity of imaging. Furthermore, the design of integrated diagnosis and treatment PA contrast agents is also a hot topic in PA molecular imaging.Conclusions and ProspectsAlthough PA imaging has made a lot of breakthroughs in recent years, it has not yet achieved a large-scale clinical application. In the next few years, the resolution, depth, speed and sensitivity of PA imaging will still be the research focus of researchers in related fields. Costs will be further reduced by using low-cost laser sources, while imaging speed and quality will meet the needs of most clinical applications thanks to technological advances in related fields. In addition, by the design of the PA contrast agent in the second near-infrared window (NIR-Ⅱ), the depth of PA imaging will further increase. We believe that through the joint efforts of researchers in related fields, PA imaging will play a more important role in the clinic practices.
ObjectiveSince the outbreak of COVID-19, many hospitals have become overloaded with patients seeking examination, resulting in an imbalance between medical staff and patients. These high concentrations of people in hospital settings not only aggravate the risk of cross-infection among patients, but also stall the public medical system. Consequently, mild and chronic conditions cannot be treated effectively, and eventually develop into serious diseases. Therefore, the use of deep learning to accurately and efficiently analyze X-ray images for diagnostic purposes is crucial in alleviating the pressure on medical institutions during epidemics. The method developed in this study accurately detects dental X-ray lesions, thus enabling patients to self-diagnose dental conditions.MethodsThe method proposed in this study employs the YOLOV5 algorithm to detect lesion areas on digital X-ray images and optimize the network model’s parameters. When hospitals and medical professionals collect and label training data, they use image normalization to enhance the images. Consequently, in combination with the network environment, parameters were adjusted into four modules in the YOLOV5 algorithm. In the Input module, Mosaic data enhancement and adaptive anchor box algorithms are used to generate the initial box. The focus component was added to the Backbone module, and a CSP structure was implemented to determine the image features. When the obtained image features are input into the Backbone module, the FPN and PAN structures are used to realize feature fusion. Subsequently, GIOU_Loss function is applied to the Head moudule, and NMS non-maximum suppression is used to generate a regression of results.Results and DiscussionsThe proposed YOLOV5-based neural network yields satisfactory training and testing results. The training algorithm produced a recall rate of 95%, accuracy rate of 95%, and F1 score of 96%. All evaluation criteria are higher than those of the target detection algorithms of SSD and Faster-RCNN (Table 1). The network converges to smoothness after loss is reduced in the training process (Fig. 6), which proves that the network successfully learns the necessary features. Thus, the difference between predicted and real values is very small, which indicates good model performance. The mAP value of network training is 0.985 (Fig. 7), which proves that the network training meets the research requirements. Finally, an observation of the visualized thermodynamic diagram reveals that the network’s region of interest matches the target detection region (Fig. 8).ConclusionsThis study proposes the use of the YOLOV5 algorithm for detecting lesions in dental X-ray images, training and testing on the dataset, modifying the network’s nominal batch size, selecting an appropriate optimizer, adjusting the weight parameters, and modifying the learning rate attenuation strategy. The model’s training results were compared with those of algorithms used in previous studies. Finally, the effect of feature extraction was analyzed after the thermodynamic diagram was visualized. The experimental results show that the algorithm model detects lesion areas with an accuracy rate of more than 95%, making it an effective autonomous diagnostic tool for patients.
ObjectiveCells are the basic units of biomass, and their morphological structures are often associated with the functional state of biomass. Therefore, the morphology of a cell is an important research topic in life science and a critical factor in clinical medical diagnosis. Quantitative phase imaging technology, as a powerful nondestructive and label-free imaging tool, provides various biological and physical properties for the quantitative evaluation of cells. Although the phase diagram of the sample provided by this technology contains information about its internal structure, the thickness and refractive index of the sample is coupled with the phase data. Decoupling the phase data is required to reconstruct a three-dimensional (3D) morphology of the sample. Dual-wavelength imaging technology is effective for single medium samples. However, this method does not work for multimedia phase objects. In response to this shortage, this study proposes a new reconstruction method based on orthogonal dual-wavelength measurements.MethodsThe 3D reconstruction method is based on three phase images from two orthogonal directions. Two of these phase images are obtained with two different wavelengths. The first step is to separate the phase shift due to different substructures. Given that the environmental liquid is a highly dispersive material relative to the cell sample, the refractive index (RI) of the environmental liquid correspondingly changes under different incident light, whereas the RI of the sample remains constant. Thus, by subtracting the two images at two different wavelengths, the physical thickness of the media adjacent to the environment (such as cytoplasm) can be determined. Next, the average RI of the cytoplasm can be extracted using the associated phase value distribution, while phase shifts due to cytoplasm and nucleus are also separated immediately. Following that, the thickness information of the nucleus for the incidence along the two directions can be obtained using a phase diagram from the orthogonal direction. Thus, the RI of the nucleus can be calculated from the nuclear phase value. The relative position of the cytoplasm and nucleus can also be determined using two orthogonal phase diagrams. The 3D morphology of the multimedia phase object is obtained by combining the physical thickness distributions of the cytoplasm and nucleus.Results and DiscussionsThe reconstructions of models with different structural characteristics are explored, including a cell with a single spherical nucleus (Fig. 2), a cell with a single saddle shape nucleus [Fig. 5(a)], and a binuclear cell with a double spherical nucleus [Fig. 6(a)]. The results of these samples [Figs. 9, 11(c), and 12(e)] are consistent with the initial model. Especially, the analytic method provides a sharp reconstruction result of the physical thickness of the cytoplasm and the entire reconstruction process takes a short time (Tables 1, 2, and 3). This study suggests the feasibility of this reconstruction method, but the actual application effect depends on many factors, such as image noise, heterogeneity of RI distribution, and calculation error in edge detection. An emphasis of the following study is to explore an efficient reconstruction algorithm suitable for experiments.ConclusionsThis study proposes a 3D morphological reconstruction method for nucleated cells based on orthogonal dual-wavelength phase images. This method requires three phase images from two orthogonal directions and is divided into two steps. First, using the high dispersion characteristics of environmental liquid and edge detection, the phases of the cytoplasm and nucleus are separated based on the independence and superposition of phase data, and the thickness of the cytoplasm is decoupled simultaneously. Then, the 3D morphology of the sample is reconstructed using another orthogonal phase diagram, RI and thickness information of the coupling nucleus, and the relative position relationship of the substructure expressed by two mutually orthogonal phase diagrams. This method collects sample information from two directions simultaneously. A small amount of data means convenient data acquisition and fast data processing. The simulation results show that the algorithm, which may provide a reference for real-time imaging of biological cells, is effective.
ObjectiveWith the increasing prevalence and blindness rate of fundus diseases, the lack of ophthalmologist resources is increasingly unable to meet the demand for medical examination. Given the shortage of ophthalmic medical staff, long waiting process for medical treatment, and challenges in remote areas, there is an irresistible trend to reduce the workload of medical staff via artificial intelligence. Several studies have applied convolutional neural network (CNN) in the classification task of fundus diseases; however with the advancement of Transformer model application, Vision Transformer (ViT) model has shown higher performance in the field of medical images. ViT models require pretraining on large datasets and are limited by the high cost of medical image acquisition. Thus, this study proposes an ensemble model. The ensemble model combines CNN (EfficientNetV2-S) and Transformer models (ViT). Compared with the existing advanced model, the proposed model can extract the features of fundus images in two completely different ways to achieve better classification results, which not only have high accuracy but also have precision and sensitivity. Specifically, it can be used to diagnose fundus diseases. This model can improve the work efficiency of the fundamental doctor if applied to the medical secondary diagnosis process, thus effectively alleviating the difficulties in diagnosis of fundus diseases caused by the shortage of ophthalmologist staff, long medical treatment process, and difficult medical treatment in remote areas.MethodsWe propose the EfficientNet-ViT ensemble model for the classification of fundus images. This model integrates the CNN and Transformer models, which adopt the EfficientNetV2-S and ViT models, respectively. First, train the EfficientNetV2-S and ViT models. Then, apply adaptive weighting data fusion technology to accomplish the complementation of the function of the two types of models. The optimal weighting factors of the EfficientNetV2-S and ViT models are calculated using the adaptive weighting algorithm and then the new model (EfficientNet-ViT) is integrated with them. After calculating the weighting factors 0.4 and 0.6, multiply the output of the ViT model by a weighting factor of 0.4, multiply the output of the EfficientNetV2-S model by a weighting factor of 0.6, and then weigh the two to obtain the final prediction result. According to clinical statistics, the current common fundamental disease in my country includes the following diseases: diabetic retinopathy (DR), age-related macular degeneration (ARMD), cataract, and myopia. These fundus diseases are the main factors that cause irreversible blindness in my country. Thus, we classify fundus images into the following five categories: normal, DR, ARMD, myopia, and cataract. Furthermore, we use three indicators, such as accuracy, precision, and specificity. The EfficientNet-ViT ensemble model can extract the features of fundus images in two completely different ways to achieve better classification results and higher accuracy. Finally, we compare the performance indicators of this model and other models. The superiority of the integrated model in the fundus classification is verified.Results and DiscussionsThe accuracy of EfficientNet-ViT ensemble model in fundus image classification reaches 92.7%, the precision is 88.3%, and the specificity reaches 98.1%. Compared with EfficientNetV2-S and ViT models, the precision of EfficientNet-ViT ensemble model improves by 0.5% and 1.6%, accuracy improves by 0.7% and 1.9%, and specificity increases by 0.6% and 0.9%, respectively (Table 3). Compared with Resnet50, Densenet121, ResNeSt-101, and EfficientNet-B0, the accuracy of the EfficientNet-ViT ensemble model increases by 5.4%, 3.2%, 2.0%, 1.4%, respectively (Table 4), showing its superiority in the fundus image classification task.ConclusionsThe EfficientNet-ViT ensemble model proposed in this study is a network model combining a CNN and a transformer. The core of the CNN is the convolution kernel, which has inductive biases, such as translation invariance and local sensitivity, and can capture local spatio-temporal information but lacks a global understanding of the image itself. Compared with the CNN, the self-attention mechanism of the transformer is not limited by local interactions and can not only mine long-distance dependencies but also perform parallel computation. This study uses the EfficientNetV2-S and ViT models to calculate the most weighted factors for the CNN and Transformer models through the adaptive weighted fusion method. The EfficientNet-ViT can extract image features in two completely different ways. Our experimental results show that the accuracy and precision of fundus image classification can be improved by integrating the two models. If applied in the process of medical auxiliary diagnosis, this model can improve the work efficiency of fundus doctors and effectively alleviate the difficulties in diagnosis of fundus diseases caused by the shortage of ophthalmic medical staff, long waiting process for medical treatment, and difficult medical treatment in remote areas in China. When more datasets are used to train the model in the future, the accuracy, precision, and sensitivity of automatic classification may be further improved to achieve better clinical results.
ObjectiveThe occurrence of numerous diseases, including cancer, cardiovascular diseases, and degenerative diseases, is closely related to the specific high expression of GSH. For instance, in A549 human lung adenocarcinoma cells, the GSH concentration was approximately an order of magnitude higher than that in normal cells. Therefore, the development of highly sensitive GSH detection and imaging approaches has crucial clinical value for the diagnosis of related diseases and a better understanding of the pathogenesis of the disease. The development of highly sensitive deep imaging approaches that can achieve GSH-specific responses in tumor tissues is still urgently needed. Photoacoustic (PA) imaging technology, as a novel biomedical imaging approach, which combines the high sensitivity of optical imaging with the deep penetration capability (up to 10 cm) of ultrasonic imaging, has been favored by the field of biomedical imaging in the last two decades. However, the characteristic molecules of many major diseases have weak optical absorption in the optical window of biological tissue (NIR-Ⅰ, 650-950 nm; NIR-Ⅱ, 950-1700 nm), resulting in the inability to generate a strong enough signal under excitation light irradiation, so that it is impossible to Realize photoacoustic imaging. Thus, developing nanoprobes with specific optical absorption properties as exogenous contrast agents for photoacoustic imaging can enhance the photoacoustic signal, thereby greatly improving the imaging contrast. Presently, various nanomaterials have been developed as exogenous contrast agents for photoacoustic imaging, including noble metal nanoprobes, carbon-based nano-2D materials, and high molecular polymers. Most of these materials are not biologically responsive and cannot specifically respond to specific substances in cells; they often lack the specific ability to recognize diseases. In this study, the authors developed a photosensitive AgBr@PLGA nanoprobe that can specifically respond to highly expressed GSH in the tumor microenvironment and proposed a tumor-specific near-infrared second region (NIR-Ⅱ) photoacoustic imaging approach.MethodsThe synthesized photosensitive AgBr@PLGA nanoprobes can be passively targeted to tumor tissue and can generate optical latent images triggered by external white light LEDs. GSH in the tumor microenvironment can reduce these optical latent images, resulting in a considerable number of silver nanoparticles that demonstrate strong light absorption and sharp improvements in photoacoustic signal in the NIR-Ⅱ region, thereby realizing specific photoacoustic imaging of tumor tissue.Results and DiscussionsThe authors characterize the morphology and optical properties of the synthesized nanoprobes, and confirm in vitro their photosensitivity and GSH response characteristics in response to externally triggered white light LEDs. The experimental findings show that the prepared nanoparticles have good biocompatibility and ultra-high sensitivity to external trigger light, and the photoacoustic signal is continuously strengthened as exposure time increases. The model demonstrated that the synthesized AgBr@PLGA nanoprobes can attain high-contrast tumor-specific imaging in vivo, demonstrating the synthesized photosensitive great application potential of nanoprobes in tumor-specific photoacoustic detection and diagnosis.ConclusionsIn this research, AgBr@PLGA nanocrystals were successfully used for ultrahigh-sensitivity and tumor-specific photoacoustic imaging through optical writing and redox chromogenic reactions. AgBr@PLGA NCs can show improved NIR-Ⅱ absorption because of the reduction of Ag nanoparticles when exposed to external trigger light after activation by GSH at the tumor site, as illustrated in Figure 2. The tumor-rich GSH content reduces the turn-on of NIR-II light absorption of AgBr@PLGA nanocrystals, enabling tumor-specific photoacoustic imaging with relatively high imaging depth, as demonstrated in Figure 5. Furthermore, this technique can accomplish contrast improvement in the tumor area by controlling the exposure time, and employing this approach can suppress unwanted background signals, such as blood signals in molecular imaging, as demonstrated in Figure 4. However, it should be noted that the response of the material at the imaging wavelength is not the same as the position of the absorption peak. We will develop photosensitive materials with strong absorption in the NIR-Ⅱ region in the following study to achieve deeper PA imaging. This study will attract more attention to the development of effective activatable PA probes for accurate biomedical imaging.
SignificanceIt is well-known that subcellular organelles are essential components of cells. Their morphological structures and dynamic characteristics directly reflect the physiological state of cells. Scientists have paid significant attention to the observation and analysis of the fine structures of subcellular organelles in living specimens.The emerging super-resolution microscopy (SRM) techniques in the early 21st century, such as structured illumination microscopy (SIM), stimulated emission depletion (STED), and single-molecule localization microscopy (SMLM), skillfully bypass the limitation of the optical diffraction limit and effectively retain the advantages of optical microscopy. SRM techniques have been widely used in monitoring subcellular organelles in living cells.ProgressThis article systematically elaborates and analyzes the super-resolution structure characteristics of subcellular organelles in living cells. First, it briefly introduces the basic principles and fundamental characteristics of the three kinds of SRM techniques, i.e., STED, SIM, and SMLM, and expounds their development status. Second, the super-resolution fine structures and dynamic characteristics of subcellular organelles, such as the nucleus, cytoskeleton, mitochondrion, and endoplasmic reticulum (ER), are presented.In 2016, Chagin et al. quantitatively measured and analyzed replication foci (RF) in mammalian cells using three-dimensional (3D) SIM. Mitchell-Jordan et al. (2012) directly imaged histone protein H3 in mammalian cells using STED to show the chromatin domain characteristics at the scale of 40-70 nm. Wombacher et al. and Lukinavicˇius et al. also employed STORM to observe the distribution of histone protein H2B in living HeLa and U2OS cells, respectively. Pelicci et al. (2020) imaged nuclear Lamin-A in intact nuclei of living cells through SPLIT-STED. Otsuka et al. (2016) captured images of different steps involved in assembling the NPC in a human cell. Lu et al. and Zhao et al. also realized the NPC super-resolution fluorescence imaging using different methods.Gustafsson et al. (2009) employed SIM to monitor the dynamic characteristics and fine structures of microtubules. Additionally, Li Dong et al. further investigated the fine structures of the cytoskeleton based on SIM. Shao et al. (2011) clearly observed that microtubules in Drosophila S2 cells showed wrapped reticular structure and were distributed sparsely in these 3D-SIM images. Zhuang Xiaowei et al. (2012) revealed the 3D ultrastructure of the microfilament skeleton using the dual-objective STORM (Fig. 2). D’Este (2015) combined the two-color STED nanoscopy with SiR-Actin to show that the periodic cytoskeleton organization is ubiquity in axons and dendrites of living hippocampal neurons. Lukinavicˇius et al. (2014) disclosed the ninefold symmetry of the centrosome and the spatial organization of the actin in the axons of rats using STED. Recently, Wang et al. (2022) proposed JSFR-SIM and followed the microtubule motion in live COS-7 cells.Additionally, Shim et al. (2012) observed the dynamic processes of mitochondrial fission/fusion through the STORM images of the mitochondrial membrane of living BS-C-1 cells. In 2020, the image data, hyperfine structures of mitochondria, and dynamic processes at different time points in living HeLa cells were attainable using STORM. Huang et al. (2018) successfully found the changes in mitochondrial crista during the fission and fusion of the mitochondrial. They identified the inter-cristae mergence in a single non-fusion mitochondrion using the Hessian-SIM system suitable for long-term super-resolution imaging of living cells (Fig. 3). Guo et al. (2018) also combined multicolor imaging technology with the newly proposed GI-SIM to observe the mitochondrial fission/fusion events at the ER-mitochondria contact sites. Wang et al. (2019) and Yang et al. (2020) accomplished the dynamic monitoring of mitochondrial crista using STED (Fig. 4). Recently, Wang et al. (2022) visualized the mitochondrial dynamics of living COS-7 cells through JSFR-SIM. A mitochondrion extended a tubulation tip, made contact with another mitochondrion, and then immediately retreated in the opposite direction.Furthermore, Shim et al. (2012) successfully realized the STORM dynamic imaging of ER membrane (Fig. 5) and expressly observed the previously obscured details of morphological changes in ER remodeling. Georgiades et al. (2017) quantitatively analyzed the length and diameter of ER tubules using STORM. Guo et al. (2018) employed GI-SIM to obtain the formation and disappearance of ER contraction sites and the reconstruction of ER tubules in living COS-7 cells. Zhu et al. (2020) recently realized the real-time STED monitoring of 3D dynamic interaction between ER and mitochondria.Finally, the development potential of combining super-resolution imaging with machine learning in exploring the fine structures of subcellular organelles is discussed.Conclusion and ProspectIt is an inevitable trend in cell image processing fields to apply deep learning algorithms in extracting information from subcellular super-resolution fluorescence images and help researchers analyze the image data. To achieve accurate and robust subcellular super-resolution image analysis, it is necessary to solve the problems of insufficient standardization of datasets and poor generalization ability of algorithm models.
ObjectiveCardiovascular is one of the major diseases that threatens human health, and the prevalence of cardiovascular disease in China continues to grow. Therefore, it is important to select an appropriate model organism to understand the development of the heart. Locust has the characteristics of easy operation, strong plasticity, and short development cycle as well as the similar gene regulation mechanism with human beings in the process of cardiac development, therefore it becomes a useful candidate for studying the cardiac function and for the pathological gene analysis. Researchers have proposed a variety of methods to evaluate the heart function of insects, such as multi-sensor electrocardiogram, atomic force microscope monitoring, and electrical stress method. However, these methods are invasive and cannot monitor the same living body continuously. Therefore, a method which can monitor the heart development and screen the phenotypic variation of insects or other model organisms non-invasively is more desiderated. Fortunately, optical coherence tomography (OCT), widely used in biomedical detection because of its noninvasiveness, real-time, and high resolution, can be used to detect the internal structures of biological tissues and other non-uniform scatterers. Therefore, it is a more suitable tool to monitor the embryonic heart development of a locust. In addition, the measurement of cardiac function parameters (such as heart rate) still needs to be calculated manually by the M-Mode diagram, which is not only time-consuming but also prone to errors. Therefore, a high efficiency automatic detection algorithm is a critical issue to be solved urgently in the high-throughput screening and phenotypic analysis of model biological pathogenic genes.MethodsUsing a locust as the model organism, in our previous works we have monitored the embryo development and screened the phenotypic variation caused by the RNAi technology. Here, a new method is proposed to automatically and quickly calculate the insect heart function parameters, such as end diastolic diameter (EDD), end systolic diameter (ESD), end diastolic area (EDA), end systolic area (ESA) and heart rate (HR). The processing flow is shown in Fig. 2. The collected 3D data are expanded in time series to obtain the M-Mode diagram of the embryo heart chamber. After gray-scale transformation of the M-Mode diagram, by a series of operations including threshold-segmentation-based regional growth, boundary recognition, morphological processing, and feature peak extraction, the parameters including HR, EDD and ESD can be obtained.Results and DiscussionsThe low-frequency noise in the original M-Mode image [Fig. 3(a)] is removed after gray-scale transformation [Fig. 3(b)], which is beneficial for the calculation by the regional growth algorithm. Then, any point selected in the fetal heart ventricle [the red dot in Fig. 3(c)] can be used as the initial seed point, and the binary regional growth result can be obtained under the specified regional growth criterion [Fig. 3(d)]. As shown in Fig. 3(d), there are burrs at the edge of the ventricle caused by the non-uniformity of the grayscale distribution, which adversely influences the accuracy in obtaining the heart beat amplitude in the next step. To solve this problem, morphological processing is introduced, which plays a good role in smoothing the cavity edge. The image after removing burrs is shown in Fig. 3(e). By counting the numbers of pixels with the logical value of 0 in A-scan and knowing the size of single pixel, the beat amplitude of the heart at different moments can be obtained [Fig. 3(f)]. As shown in Fig. 3(g), the HR, EDD, and ESD cardiac parameters can be calculated after the extreme points are found by the peak extraction algorithm. If the original image is changed from the M-Mode image to the B-scan image of the cross section of the embryonic heart, the maximum EDA and the minimum ESA of the locust embryonic heart can be calculated according to the steps in section 2.2, as shown in Fig. 6. Therefore, one can automatically detect and quantitatively analyze the heart function parameters of insect embryos by the proposed algorithm.ConclusionsIn the field of heart development and mechanism of heart disease, OCT has been successfully applied to detect the heart function of model organisms such as insects due to its advantages of noninvasiveness, real-time, and high resolution. However, the detection algorithm still has some problems, such as low efficiency, high requirements on image quality, and inaccuracy of measurement, especially it is not suitable for the detection under a large sample size. In this paper, we propose a high speed automatic detection and quantitative analysis algorithm of insect cardiac function parameters by OCT. The position of the seed point is determined through human-computer interaction, and a series of processing such as automatic image segmentation and target region division are performed on the OCT M-Mode image of the insect heart. The proposed algorithm can quickly and accurately measure the cardiac function parameters including the end diastolic diameter, end systolic diameter, end diastolic area, end systolic area, and heart rate. This method can improve the screening and analysis efficiency of pathogenic genes in high-throughput biological samples and has important applicable value in the research of cardiovascular disease using insects as model organisms.
ObjectiveDeveloping a fiber probe with a high lateral resolution, long depth of focus, long working distance, and uniform axial light intensity is essential for endoscopic optical coherence tomography (OCT). Traditional beam shaping components, such as axicon lens and binary-phase mask, were minimized and adapted to miniature fiber probes for optimized output beams, but with a short working distance and reduced depth of focus gain compared to their bulk-optic counterparts. Alternatively, pure fiber techniques that simply splice fibers in series were proposed and demonstrated a significant enhancement in the imaging quality. The first demonstration of the concept suggested a phase mask consisting of a short section of overfilled graded-index fiber (Lorenser, 2012). However, the most recent progress indicated that using a step-index large core fiber as a coaxially focused multimode beam generator (Yin, 2017) or a high-efficient fiber-based filter (Ding, 2018) would be advantageous in terms of easy fabrication and increased depth of focus gain. However, full optimization of such probes is not straightforward, where the difficulty originates from the complexity of the light field by multimode interference and the arrangement flexibility of fiber components. Therefore, this study presents systematic research on the optimization of large core fiber probes. We discuss key design considerations for selecting fiber optics for mode excitation, number of modes, beam expansion method, and selecting the lens-free mode or spatial filter mode. We hope that our findings can be essential in designing the ultrathin fiber probe with improved performance for OCT imaging.MethodsA unified equation quantifying the depth of focus gain was first deduced by comparing the Gaussian beam with the same minimal beam diameter. Then, the fiber mode theory was applied to demonstrate the light field manipulated by the large core fiber. The tunning length of the large core fiber was determined by its re-imaging property and modal dispersion. According to the relative position of the large core fiber to the pupil of the objective, the working modes of the probe were classified into two catalogs. Consequently, proximate equations of the output light field were deduced for the two working modes. Then, the optimization goals and restrictions were established in terms of the depth of focus gain, lateral resolution, uniformity of axial light intensity, working distance, and sidelobes. The fast simulation method based on the mode expansion was applied to search for the probe parameters according to the established goals and restrictions. We obtained the preferable structure and the maximum achievable performance of the large-core-fiber-based probe by comparing the optimized results under different working modes, beam expansion methods, and the number of modes. The fiber probe with optimized parameters was fabricated and interfaced to a swept-source OCT system. A conventional probe with the same minimal beam diameter was also fabricated for comparison. The same region of fresh lemon was scanned with a translational stage and imaged by the two probes to confirm whether the improved specifications led to corresponding enhancement in the imaging quality.Results and DiscussionsSimilar to the binary phase filter that regulates the output beam by encoding the phases of annular zones on the aperture, the large core fiber can also adjust modal phases independently for the output beam manipulation. Additionally, the depth of focus, working distance, and lateral resolution are expected to increase with introduced higher modes. Although sidelobes become more significant with increased depth of focus, they can be mitigated by optimizing the modal power distribution. The large-core-fiber-based probe has less stringency on fabrication than the fiber phase mask. With a length tolerance of -28-+ 20 μm (Fig. 8), it is achievable for a commercial off-the-shelf fiber processing platform. The large-core-fiber-based probe features axially uniform light intensity compared with the coaxially focused multimode beam generator (Figs. 10 and 12). For the mode excitation device, we find that the graded-index fiber is superior to the tapered fiber in terms of a more robust splicing point. For the dual-mode interference, the amplitude ratio of the fiber mode can be tuned in the range of 0.2-0.3. For multimode interference, the graded-index fiber can be used with the no core fiber for efficient higher-order mode excitation (Fig. 12). For the working modes of the probe, we conclude that the spatial filter mode is advantageous in a larger depth of focus gain. Additionally, a longer working distance is attainable with beam relay optics (Table 4). The modal dispersion is the ultimate limit on the performance of the large-core-fiber-based probe. We confirmed that the maximum DOF gain of the probe was 3.8.ConclusionsThis study systematically investigates the optimization method of a large-core-fiber-based probe. By comparing the lateral resolution, working distance, and focal depth gain of various probe designs, we believe that the spatial filter design with beam relay optics and higher-order modes is beneficial for the probe performance. In addition to OCT imaging, the principle of the framework can be applied to optimize the output beam in laser scanning and photoacoustic imaging systems. Some presented elements of the work can also provide technical implications for non-imaging applications, such as fabrication of laser-fiber couplers and optical tweezers.
ObjectiveReducing excitation intensity or exposure time is employed to decrease the phototoxicity and photobleaching in structured illumination microscopy (SIM). However, the raw images obtained under this condition have a low signal-to-noise ratio, resulting in an error estimation of parameters and reconstruction artifacts. To improve the accuracy of parameter evaluation, some modified parameter evaluation algorithms have been proposed, including the prefiltering approach and iterative and noniterative parameter evaluation approaches. However, these approaches can only enhance the accuracy of the estimated parameters, but do not demonstrate how precise they are. In other words, these algorithms cannot assess whether there is a considerable deviation from the true value. However, to suppress the reconstruction artifacts in the reconstructed image, some reconstruction algorithms have been proposed, such as spectrum filtering, TV-SIM, and Hessian-SIM. These approaches usually reconstruct the super-resolution (SR) SIM image at the beginning, and then remove the artifacts. However, this reconstruction process will change the Poisson-Gaussian noise distribution in the images.To address the above two issues, we first proposed a parameter evaluation approach based on empirical mode decomposition (EMD) in this research, which can precisely evaluate the initial phase and modulation depth. Measured with the proposed dispersion index, the accuracy of the estimated parameter can be given synchronously. Next, a denoising algorithm based on similar blocks was employed before SIM reconstruction in this study, which maintains the illumination pattern while suppressing the noise in the raw images. This predenoise process before reconstruction can enhance the accuracy of parameter evaluation and remove the artifacts.MethodsThis study employs empirical modal decomposition (EMD) to smooth out the frequency distribution of the estimated initial phase and modulation depth. The specific process is as follows. First, the distribution curve of the parameter estimate was decomposed with each inherent mode function component. Next, only the mode component below 4 was accumulated, which can be superimposed as a smooth fitting curve. Finally, we evaluated the initial phase and modulation depth by this superimposed smooth curve.The dispersion index was proposed as a quantitative index that can be used to measure the evaluated initial phase and modulation depth accuracy in this study. This index primarily characterized the degree of concentration of the curve from the global and local aspects.This study also suggested a new process of denoising the raw images first and then conducting SR reconstruction, which can enhance the accuracy of the parameter estimation and reduce the artifacts. First, we obtained the average of the raw images and concatenated it with the raw images. Next, a VST transformation was performed on the concatenated images, followed by a VBM3D-based denoising process. Finally, the denoised findings were subjected to a VST inverse transformation. After the complete denoising process, we performed the subsequent SR reconstruction.Results and DiscussionsWe first imaged actin with 20 groups for comparison experiments. Each group contained 9 raw images, with three phases in every three directions, for a total of 180 images with 486 ms. These 20 groups of raw images can be directly reconstructed into 20 SR images by the Wiener reconstruction algorithm. Thereafter, we averaged the 20 SR images to one image that serves as the ground truth.For the parameter evaluation, the frequency distribution of the parameter estimates is ideally similar to the shape of the impulse function [Fig. 1(b)]. However, because of the effect of noise, the actual distribution is a smooth curve with local jitter [blue solid line in Fig. 1(c)]. The EMD algorithm was then employed to fit this curve to make parameter estimation easier [Fig. 1(c) red dashed line].To evaluate the accuracy of the estimated initial phase and modulation depth, we proposed a dispersion index and confirmed its effectiveness. First, we averaged the first 1, 3, 5, 7, 9, and 11 groups of the raw images to obtain six groups of images with various SNR. These six group images were employed as experimental data for the evaluation of the dispersion index. The experiment reveals that with the enhancement of the raw images' SNR, the EMD curve [Fig. 4(d)-(e) red dashed line] is more similar to the shape of the impulse function. Moreover, the dispersion index gradually decreases [Fig. 4(c)]. Therefore, the dispersion index characterized the relationship between the SNR of raw images and parameter evaluation accuracy.The two benefits of the proposed denoising algorithm are also confirmed. First, we found that the EMD curve is more concentrated [Fig. 6(b)-(c)] and the dispersion index is low (Table 1 and Table 2), implying that the calculated parameters are more accurate when using the VST-VBM3D denoising algorithm. In the meantime, we discovered that the artifacts are suppressed [Fig. 7(d)] and the PSNR and SSIM values were high (Table 3) when using the proposed denoising algorithm.ConclusionsThis study proposes a parameter evaluation algorithm based on the EMD algorithm and corresponding dispersion index, which can accurately evaluate the initial phase and modulation depth from the raw images and simultaneously analyze the evaluation accuracy. Simultaneously, this study also proposes a VST-VBM3D-based denoising algorithm for SIM raw images, which may suppress the noise in the raw images before the SR reconstruction procedure. This predenoise approach not only enhances the accuracy of parameter evaluation but also reduces the artifacts in reconstructed SR images.
ObjectiveThe propagation of light in turbid media is affected by the optical parameters of the media including absorption coefficient (μa), scattering coefficient (μs), isotropic coefficient (g), and refractive index (n). These optical parameters are related to the chemical properties, the internal structures, the physical properties of the media, and the boundary difference and speed of light transmission, including the shape, size, and concentration of different scattering components in the turbid media. By measuring the optical parameters of the turbid media, the material properties, physiological states and pathological changes can be determined, which is very important in various applications including biomedical diagnosis and food safety inspection. However, there is a lack of algorithms that can be simultaneously used to identify these four parameters (μa,μs,g, and n) because the measurement instruments cannot be easily installed. To solve this problem, a method based on a residual neural network is proposed here to determine the four parameters of the turbid media from the diffuse reflection light intensity profiles.MethodsFirst, the diffuse reflection light intensity profiles under different incident excitation light angles are obtained through the Monte Carlo simulation. The incident light spot diameter and the divergence angle are considered in the simulation process. Second, the diffuse light intensities excited under multiple angles are used to enhance the information richness. Third, a residual neural network is used to establish the machine learning mapping model between the diffuse light intensity profiles and the optical parameters of the turbid media, and the prediction of optical parameters is realized. The extracted light intensity values along the long axis are used as the input of the residual neural network, and the output is the optical parameters. Before training and testing, noise is added to the diffuse reflection data in order to simulate the optical measurements under real conditions. The input data is normalized to ensure the consistency of data range and make the network converge quickly.Results and DiscussionsIn the Monte Carlo simulation, different incident angles are initialized. Seven incident angles ( 24°, 30°, 36°, 42°, 48°, 54°, and 60° ) are applied in this work (Fig. 4). The position projected by each photon on the medium surface is initialized as (x′, y′, z′) and the photon directional cosine is set as (μx,μy,-μz). The diffuse light intensity profiles under different excitation light angles are validated to be linearly independent (Fig. 6). Thus they may provide extra effective independent constraints for the estimation of the four optical parameters. The concept of using more diffuse reflection light intensity profiles to enhance data richness is further proved by the full rank of diffuse reflective light intensity vectors along the long axis. The relative error decreases with the increase of the number of diffuse reflection light intensity profiles used here (Fig. 7). When only the diffuse reflection light intensity at one angle is used to identify the optical parameters of the media, the errors can be several times larger than those when the 7 sets of diffuse reflection light intensity profiles are used. The recognition errors of the four optical parameters have little change when the SNR is changed in the range of 40 dB-80 dB. The results show that the prediction errors for the four optical parameters (μa,μs,g, and n) are 8.6%, 4.6%, 1.7%, and 0.9%, respectively, when the noise level is 40 dB. Compared with the existing prediction methods, the proposed residual neural network method has high prediction accuracy and short computation time.ConclusionsA method based on a residual neural network is proposed to estimate the anisotropic coefficient, absorption coefficient, scattering coefficient, and refractive index of a turbid medium. The diffuse light intensities excited under multiple angles are proved to be effective for enhancing the information richness and improving the estimation accuracy of optical parameters. The incident light spot diameter and the divergence angle are considered, and the different levels of noise are added to the diffuse light intensity signals and the generalization ability and anti-noise performance of the network are improved. The results show that the proposed method can estimate the anisotropic coefficient, absorption coefficient, scattering coefficient, and refractive index of the turbid medium accurately with a high noise level and a high efficiency. The diffuse light intensities under seven angels are enough for the determination of the four optical parameters. This work is expected to be useful for various applications including biomedical diagnosis, food safety inspection, and material property detection.
ObjectivePhotoacoustic imaging (PAI) has been a fast-growing biomedical imaging modality in recent years. Absorbers are imaged in biological tissue by detecting laser-induced ultrasound waves via PAI. This provides hemodynamic information for the diagnosis of tumors, strokes, and other critical diseases. A typical photoacoustic microscope uses piezoelectric ultrasonic transducers to detect the photoacoustic signals. However, owing to the tradeoff between sensitivity and sensor size, building a miniaturized photoacoustic microscope with superior imaging capability is highly challenging. Therefore, the applications of PAI in wearable and endoscopic applications are limited. Our research group has developed a new optical ultrasonic sensor using a self-heterodyne fiber laser as the acoustically sensitive element. However, the laser, optical amplifier, photodetector, and signal demodulator may all cause noise and limit the detection capability. This study analyzes the noise characteristics and constructs a high-performance fiber-based photoacoustic microscope. This imaging probe can be used as a gastrointestinal endoscope for tumor screening or as a head-mounted microscope for brain imaging in a free-moving state.MethodsWe exploited a compact fiber laser as the ultrasound sensor. Ultrasound waves can deform the sensor and induce a change in the lasing frequency. To measure the acoustic response, we used both x- and y-polarized light and beat them at the photodetector to generate a radio-frequency. The variation in lasing frequency was then measured at radio frequency using modern electronics with high resolution. Here, we theoretically analyzed the noise of the fiber sensor, optical amplifier, photodetector, and signal demodulation acquisition module in the ultrasonic detection system. By measuring the noise n0,beat signal power Prf,and frequency noise Δfnoise with different input optical powers, we examined the dependence of the noise on the input power. Further, we implemented a photoacoustic microscope using an optical sensor for ultrasound detection and imaged the blood vessels in a biological sample. The signal-to-noise ratios (SNRs) were also measured while varying the input power of the sensing light.Results and DiscussionsFirst, we calculated the noise levels of the fiber laser and optical amplifier, shot and thermal noises of the photodetector, and the noise of the signal demodulation acquisition module (Fig. 2). We found that when the input optical power is less than 8.5 mW, the noise from the data-acquisition system accounts for a large proportion of the total system noise and has a main contribution to the noise; when the input optical power exceeds 8.5 mW, the noise of laser and optical amplifier dominates. We then measured the system noise n0,beat signal power Prf,and image frequency noise Δfnoise as the functions of input optical power (Fig 3). When the input optical power increases to more than 10 mW, the frequency noise Δfnoise approaches its minimum and the root-mean-square of Δfnoise is ~44 kHz. In photoacoustic microscopy, the optical ultrasound sensor was used to detect laser-induced ultrasound waves. We imaged a mouse ear in vivo with different input powers (Fig 5). When the input optical power is 1.7 mW, the peak-to-peak frequency noise is 185 kHz. When the input optical power increases to 15.7 mW, the noise is reduced to 110 kHz and the imaging SNR is enhanced by 4.5 dB.ConclusionsThis paper studies the noise characteristics of a laser-based optical ultrasound sensor. We determine the dependence of noise n0 of the optical fiber ultrasonic sensor system and that of the beat signal frequency noise Δfnoise on the input optical power Prf of the signal light. By increasing Prf,the frequency noise can be considerably reduced, yielding an enhancement in the SNR. The frequency fluctuation is reduced from 185 to 110 kHz when the input power is increased to 15.7 mW. The corresponding noise equivalent pressure (NEP) is reduced from 32.9 to 19.5 Pa, and the imaging SNR is enhanced by 4.5 dB.Optical fiber is thin, flexible, and suitable for both endoscopy and wearable instrumentation. This study demonstrates that optical fiber technology opens new possibilities to implement small high-performance photoacoustic imaging modalities. Here, we have considerably improved the sensitivity of the optical ultrasound sensor, thus providing better imaging results. With the improved sensor, we aim to implement a photoacoustic endoscope for gastrointestinal cancer diagnosis and a head-mounted photoacoustic microscope for free-state neuroimaging.
ObjectiveOptical coherence tomography (OCT) imaging shows great potential in clinical practice because of its noninvasive nature. However, two critical issues affect the diagnostic capability of OCT imaging. The first problem is that the interferential nature of OCT imaging produces interference noise, which reduces contrast and obfuscates fine structural features. The second problem is caused by the low spatial sampling rate of OCT. In fact, in clinical diagnosis, the use of a lower spatial sampling rate is a method to achieve a wide field of vision and reduce the impact of unconscious movement. Therefore, most OCT images obtained in reality are not optimal in terms of signal-to-noise ratio and spatial sampling rate. There are significant differences in the texture and brightness of the retinal layer in patients, as well as in the shape and size of the lesion area, so traditional models may not be able to reliably reconstruct the pathological structure. To obtain high peak signal-to-noise ratio (PSNR) and high-resolution B-scan OCT images, it is necessary to develop sufficient methods for super-resolution reconstruction of OCT images. In this paper, an improved OCT super-resolution image reconstruction network structure (PPECA-SRGAN) was proposed.MethodsIn this paper, a PPECA-SRGAN network based on generative adversarial network (GAN) was proposed. The network model includes a generator and a discriminator. A PA module was added between the residual blocks of the generator to increase the feature extraction capability of OCT retinal image reconstruction. In addition, a PECA module was added to the discriminator, which is an improvement of the pyramid split attention network (PSANet) and can fully capture the spatial information of multi-scale feature maps. First, we used two data sets to test a training set of 1000 images and a test set of 50 images, respectively. The data set was imported into the preprocessing module, and the low-resolution image was obtained through four down-sampling processes. Then, the generator was used to train the model to generate high-resolution images from low-resolution images. When the discriminator could not distinguish the authenticity of the images, it indicated that the generation network generated high-resolution images. Finally, the image quality was evaluated using the structural similarity index measure (SSIM) and PSNR.Results and DiscussionsThe super-resolution index evaluation results of PPECA-SRGAN and the other three models were compared, as well as the final reconstruction effect images. In general, PPECA-SRGAN’s reconstruction effect was better than SRResNet; however, for the restoration of the image details, the image quality of the PPECA-SRGAN network reconstruction was more in line with the satisfaction degree of human vision. Compared with SRResNet, SRGAN, and ESRGAN, the SSIM indexes of PPECA-SRGAN were 0.090, 0.028, and 0.016 higher and the PSNR indexes were 2.15 dB, 0.71 dB, and 0.47 dB higher, respectively. The good reconstruction effect of PPECA-SRGAN was due to the addition of the attention mechanism called path aggregation network (PANet) and the proposed attention mechanism named PECA, both enhancing the capture of OCT retinal image features and the reconstruction of details. The PECA module was composed of pyramid splitting and extracting features, with the use of ECANet to fuse multi-scale information. PANet can effectively reduce image noise, such as compression artifacts. This makes our model better than the SRGAN network and other traditional networks. Therefore, the application of the proposed model in OCT image super-resolution reconstruction has been verified, and its performance has been improved compared with other reconstruction algorithms.ConclusionsThe PPECA-SRGAN network structure proposed in this paper is an improved model of the SRGAN network for super-resolution reconstruction of retinal OCT B-scan images. We conducted training and verification on MICCAI RETOUCH data set and data collected by Wenzhou Medical University to solve the problems of low-resolution and few details of images collected by OCT. We used advanced GAN to improve the super-resolution reconstruction of OCT images, and the SRGAN network was improved due to the difference in reconstruction between medical images and natural images. Firstly, a PANet module was added between the residual blocks of the generator to extract multi-scale feature relations by pyramid structure and suppress unnecessary artifacts. Then, the PECA module was inserted into the discriminator to effectively combine spatial and channel attentions to learn more image details for the discriminator and obtain richer image pair feature information. The experimental results show that this model is effective and stable in improving the resolution of medical images. Compared with SRResNet, SRGAN, and ESRGAN, the PSNR and SSIM indexes of the reconstructed images were improved by about 3.5% and 5.6%, respectively. In clinical diagnosis, the proposed algorithm can overcome the inherent limitations of low-resolution imaging systems and reconstruct various details lost in the process of image acquisition; the algorithm is easy to integrate and implement. In the future, if higher-quality data sets and lighter algorithms can be obtained, it is possible to further improve the quality of super-resolution reconstruction medical images and make them more applicable in clinical practice.
SignificanceTo minimize the physical damage, phototoxicity, and photobleaching of the biological samples, microscopic imaging methods for the visualization of cells and tissues need to have the ability of noncontract and fast measuring of the three-dimensional (3D) sample information. Far-field optical microscopy, which has been widely applied for biomedical imaging, is one of the most direct and indispensable ways of capturing the dynamic 3D architecture of biological samples. In the optical imaging system, both the intensity and phase distribution of the illumination light field will be quantitatively modulated by the sample and finally transmitted to the detector plane. The demodulation of all the obtained information enables quantitative reconstructions of the samples’ 3D spatial structure, morphology profile, and refractive index distribution. However, the existing photon detectors are only sensitive to the intensity distribution of the input light signal. The phase of the light field, which cannot be directly measured by the detector, can be quantitatively coded and decoded from the two-dimensional intensity distribution of the interference pattern using the interference characteristic of light. These basic ideas enable, in principle, fast 3D imaging, tomography, and quantitative phase-contrast imaging and hence benefit the visualization of the dynamic structural and biophysical characteristics of the samples. Furthermore, the details of the fine structures inside the sample can be obtained with improved imaging performance through super-resolution imaging and nonscanning 3D imaging, which are enabled by fluorescent self-interference imaging techniques. All these potential advantages in biological imaging have promoted the rapid development of interference microscopic techniques in the past decades. The interference imaging methods are classified according to the coherence properties of the light source used. The interference microscopic techniques using different light sources enable quantitative phase-contrast imaging, nonscanning 3D imaging, and tomographic imaging to practically benefit the structural and functional visualization of 3D complex biological samples. The advantages, limitations, and potential applications of different interference imaging techniques are shown in Table 1.ProgressDifferent optical systems and numerical methods have been designed to improve the spatial resolution, imaging speed, signal-to-noise ratio, and robustness to extend the application and modalities of the interference microscopic techniques. Among them, research is mainly focused on the applications of digital holography in quantitative phase-contrast imaging (Fig. 4) and nonscanning 3D imaging (Fig. 6) of the sample. Parallel phase shifting (Fig. 8) and compressive sensing (Fig. 9) methods have been combined with digital holography to improve temporal and spatial resolution. Digital holography’s large field-of-view, high speed, high resolution, and multidimensional imaging abilities have benefited both functional (Fig. 5) and structural (Fig. 7) imaging of biological samples. With optical sectioning imaging ability and less speckle noise, partially coherent digital holography has been applied for high accuracy phase-contrast imaging of cells (Fig. 10) and, more importantly, for the visualization of the structure behind the tissues (Fig. 11). Because of its 3D tomographic imaging ability, optical coherence tomography (OCT) has become one of the most important tools for ophthalmic imaging (Fig. 13). With extended imaging modalities, polarization sensitive OCT has provided proof-of-principle results in the diagnosis of bronchial disease (Fig. 14). Incoherent holography can considerably improve the temporal resolution of the existing 3D laser scanning fluorescence microscope. Nonscanning 3D imaging of the fluorescence sample has been demonstrated (Fig. 15) with inherent super resolution (Figs. 18 and 19). While some of the major limitations of incoherent holography, such as the low axial resolution, have been addressed and improved (Fig. 16), the potential of this technique for high-resolution, high-speed 3D fluorescence imaging is still being explored. Successes have been achieved, e.g., by optimizing the 3D imaging performance of fluorescence holography via computational adaptive optics (Fig. 17). In localization-based super-resolution microscopy, the basic idea of interference microscopy has also been used as a point spread function modulation method. Therefore, the system’s 3D resolution and imaging depth have been improved (Figs. 21 and 22).Conclusions and ProspectsIn this paper, we have reviewed the basic principles, recent progresses, advantages, limitations, applications, and potential future directions of the techniques. The system’s 3D spatial resolution, imaging speed, and signal-to-noise ratio have been considerably improved during the past decades. Based on the multidimensional (3D spatial+ phase) imaging ability of interference microscopy, the applications of the methods for the structural and functional imaging of biological samples have been demonstrated. Further, the imaging modalities have been extended to provide even more data dimensions by combing the interference microscopic techniques such as OCT with polarization imaging method. In conclusion, in interference microscopy, the concurrently obtained structural and functional information of the sample is important for understanding the biological and biophysical mechanisms of the life processes. Interference microscopic techniques have benefited specific research in biological society by providing a powerful 3D imaging tool for both coherent and incoherent light sources. While several efforts have been made to improve system spatial resolution, another important direction in the future is to further develop functional imaging methods by exploring the potential of superior multidimensional data acquisition ability.
ObjectivePhotoacoustic imaging (PAI) is an emerging biomedical imaging technique with a high contrast of optical imaging and high resolution and deep penetration of acoustic imaging, which has shown broad application prospects in the field of clinical disease diagnosis. An important implementation for realizing PAI is using acoustic-resolution photoacoustic microscopy (AR-PAM). Conventional light illumination methods have the problem of uneven light distribution, optical thermal noise, large energy loss, and decreased imaging sensitivity. Additionally, for the samples of complex biological tissues with irregular shapes, such as tumor and brain tissues, single-sided illumination methods have imaging limitations such as incomplete coverage of the target area and difficulty in obtaining accurate deep tissue information. In this paper, we report a dual-sided illumination method for AR-PAM. When compared with conventional methods, this method has higher imaging contrast in complex biological samples and can more accurately present the complete boundary of sample tissue. More comprehensive information was obtained, demonstrating the method’s promising potential in both clinical and preclinical research.MethodsA polarization splitter was used in this study to divide the laser beam into two beams and they were coupled into the multimode optical fiber through fiber couplers. After being shaped with a planoconvex lens on both sides of an imaging probe, the emitted beams were irradiated to the imaging sample at a 45° angle. A high-frequency ultrasonic transducer received the photoacoustic signals generated by the sample. First, the feasibility of imaging was verified by creating two phantoms mimicking blood vessels at different depths. The imaging of popliteal lymph nodes, brain vasculature, and tumors in living mice with two illumination methods was then performed and compared, and their imaging performance with dual-sided illumination was more excellent than that with single-sided illumination method, proving the advantages of dual-sided illumination method in PAI of complex biological samples.Results and DiscussionsWhen the PAI results of the two tissue phantoms under different illumination schemes are compared, the overall signal-to-noise ratio and contrast of the images in the dual-sided illumination method are found to be better than those in the single-sided illumination method and more complete contour and depth information can be obtained for the imaging of complex samples (Fig. 3). In in vivo imaging experiments, the advantages of dual-sided illumination in improving imaging quality are also verified. Through imaging of indocyanine green traced (ICG-traced) mouse popliteal lymph nodes, the signal intensity of lymph nodes using dual-sided illumination method was approximately three times higher than that using the single-sided illumination method (Fig. 4). Noninvasive imaging of cerebral cortical blood vessels showed that the dual-sided illumination method can present more abundant microvessels in the marginal region with higher contrast (Fig. 5). Unlabeled in vivo imaging of mouse tumors was performed to evaluate the differences in peripheral vascular imaging between the two illumination methods, and the results showed that the blood vessels observed in the same area using the dual-sided illumination method were more abundant and tumor nourishing vessels were visible (Fig. 6). In addition, three-dimensional reconstruction of the tumor image showed that the dual-sided illumination method can image tumor edges more accurately and completely.ConclusionsIn this study, the imaging quality is improved by reconfiguring the light illumination method of AR-PAM from single-sided illumination to dual-sided illumination to achieve the homogeneous coverage of a laser beam for imaging complex biological tissues. The results show that the dual-sided illumination method improves contrast and signal-to-noise ratio for PAI in complex biological samples such as tissue phantoms, popliteal lymph nodes, brain vasculature, and tumors. Our study provides a new method for photoacoustic microscopy and has the potential to improve diagnosis accuracy in clinical and preclinical practices.